You might’ve found it challenging to tailor them to your specific use cases - not anymore!
Introducing MOFA-FLEX: a flexible, modular factor analysis framework designed for customizable modeling across diverse multi-omics data scenarios. 1/n

You might’ve found it challenging to tailor them to your specific use cases - not anymore!
Introducing MOFA-FLEX: a flexible, modular factor analysis framework designed for customizable modeling across diverse multi-omics data scenarios. 1/n

Bonus: robust, efficient interpretation of syntax
Great collab with @jengreitz.bsky.social lab.

Bonus: robust, efficient interpretation of syntax
Great collab with @jengreitz.bsky.social lab.
🧵👇 (1/n)
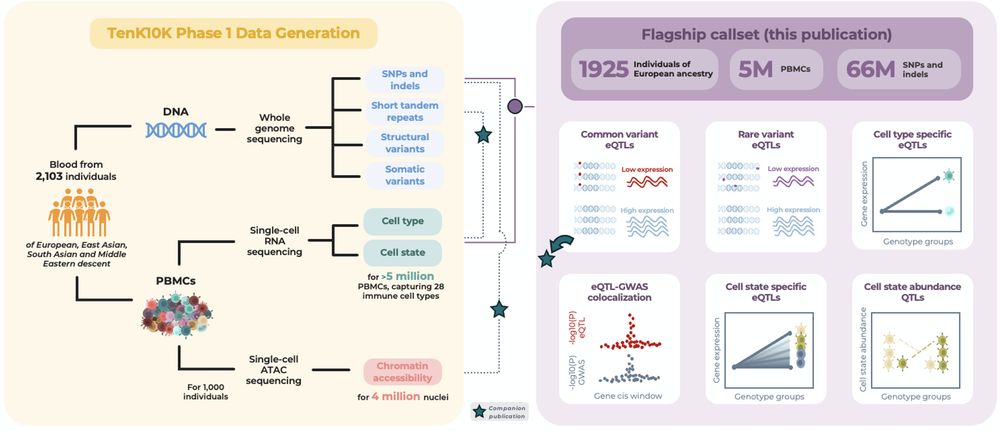
🧵👇 (1/n)
We have an exciting opportunity to join our team at GSK for a 6-9 months internship, working on an ambitious cross-department research project. Apply before March 14th!
www.linkedin.com/jobs/view/41...
We have an exciting opportunity to join our team at GSK for a 6-9 months internship, working on an ambitious cross-department research project. Apply before March 14th!
www.linkedin.com/jobs/view/41...

www.biorxiv.org/content/10.1...
Huge congrats to Anusri! This was quite a slog (for both of us) but we r very proud of this one! It is a long read but worth it IMHO. Methods r in the supp. materials. Bluetorial coming soon below 1/
www.biorxiv.org/content/10.1...
Huge congrats to Anusri! This was quite a slog (for both of us) but we r very proud of this one! It is a long read but worth it IMHO. Methods r in the supp. materials. Bluetorial coming soon below 1/
Do these studies find the most IMPORTANT genes? If not, how DO they rank genes?
Here we present a surprising result: these studies actually test for SPECIFICITY! A 🧵on what this means... (🧪🧬)
www.biorxiv.org/content/10.1...

Do these studies find the most IMPORTANT genes? If not, how DO they rank genes?
Here we present a surprising result: these studies actually test for SPECIFICITY! A 🧵on what this means... (🧪🧬)
www.biorxiv.org/content/10.1...


The team is rapidly deploying fixes and new software to adapt. More servers in the mail.
The team is rapidly deploying fixes and new software to adapt. More servers in the mail.
Haven't heard of EMBL-EBI? Take a look at what we’re working on. www.ebi.ac.uk/about/our-im...

Haven't heard of EMBL-EBI? Take a look at what we’re working on. www.ebi.ac.uk/about/our-im...
We usually find that most fine-mapped variants do not fall within coding or regulatory regions. Is it a limitation of epigenomics or a limitation of fine-mapping? Please share your thoughts!
We usually find that most fine-mapped variants do not fall within coding or regulatory regions. Is it a limitation of epigenomics or a limitation of fine-mapping? Please share your thoughts!
New preprint from a collaboration led by Grace Bower and Evgeny Kvon.
doi.org/10.1101/2024...
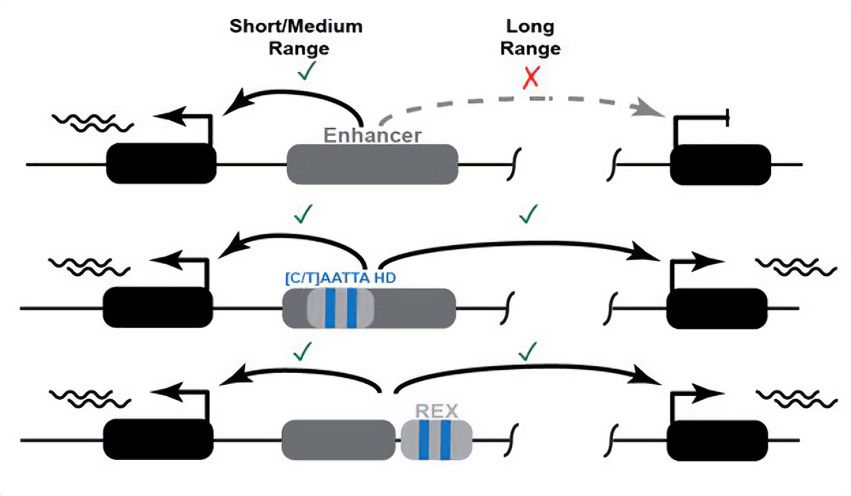
New preprint from a collaboration led by Grace Bower and Evgeny Kvon.
doi.org/10.1101/2024...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...

www.gov.uk/government/n...
www.gov.uk/government/n...


Facilitates genomic liftover using existing Bioc tools:
ranges |> easylift("hg38")
Abdullah is one of the group that signed up for #tidyomics open challenges: github.com/tidyomics

Facilitates genomic liftover using existing Bioc tools:
ranges |> easylift("hg38")
Abdullah is one of the group that signed up for #tidyomics open challenges: github.com/tidyomics
doi.org/10.1038/s415...

doi.org/10.1038/s415...

