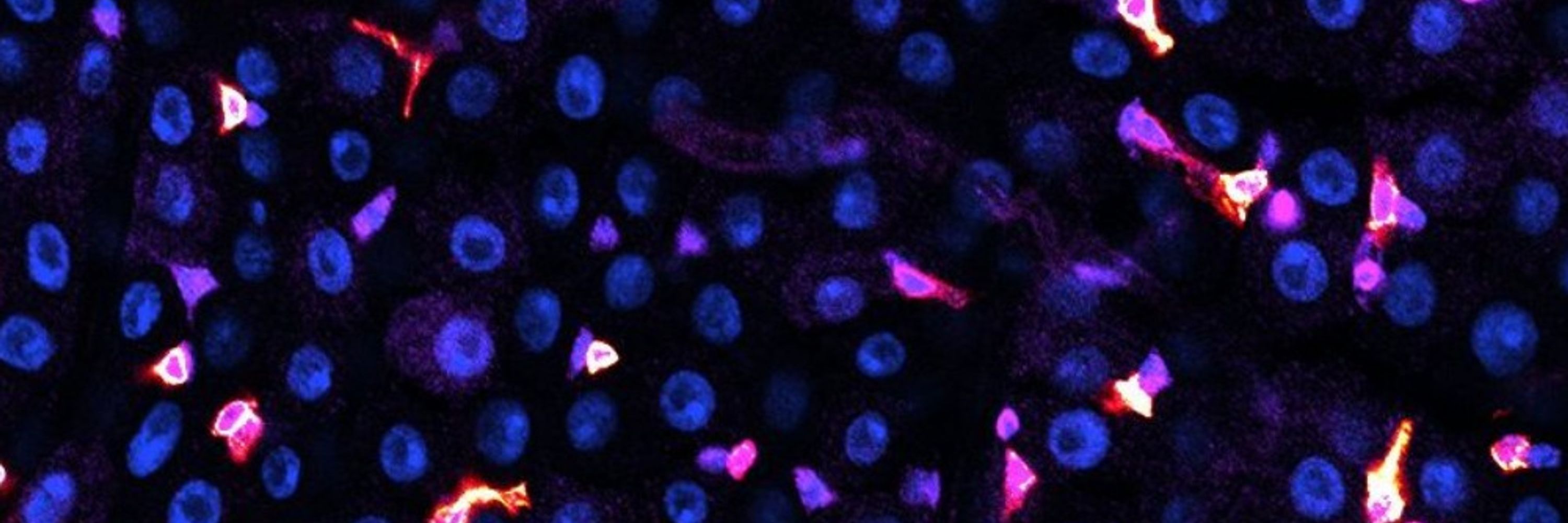
We asked: How can we make CRISPR knockouts in vivo more efficient? And set up an assay that enables detection of CRISPR cutting with 100s-1000s of sgRNAs over entire chromosome arms in living animals.
Grab a ☕ and let’s dive in. 🧵🧪 1/n
#CRISPR #geneediting
Multiplexed Cas12a editing plasmids here: www.addgene.org/browse/artic...
Multiplexed Cas12a editing plasmids here: www.addgene.org/browse/artic...
www.nature.com/articles/s41...
#CRISPR #geneediting #Drosophila 🧪🧬✂️🔬🪰
Summary 🧵 below.

www.nature.com/articles/s41...
#CRISPR #geneediting #Drosophila 🧪🧬✂️🔬🪰
Summary 🧵 below.
📄 www.science.org/doi/10.1126/...
🌐 search.foldseek.com/foldmason
💾 github.com/steineggerla...

📄 www.science.org/doi/10.1126/...
🌐 search.foldseek.com/foldmason
💾 github.com/steineggerla...
Background: The Cancer Dependency Map from @depmap.org is a fantastic resource that characterises genetic dependencies at genome-wide scale across ~1,000 cancer cell lines. 1/9
Background: The Cancer Dependency Map from @depmap.org is a fantastic resource that characterises genetic dependencies at genome-wide scale across ~1,000 cancer cell lines. 1/9




shop.vbc.ac.at/vdrc_store/v...
www.addgene.org/browse/artic...
www.plasmids.eu
shop.vbc.ac.at/vdrc_store/v...
www.addgene.org/browse/artic...
www.plasmids.eu


www.nature.com/articles/s41...
#CRISPR #geneediting #Drosophila 🧪🧬✂️🔬🪰
Summary 🧵 below.

www.nature.com/articles/s41...
#CRISPR #geneediting #Drosophila 🧪🧬✂️🔬🪰
Summary 🧵 below.
The alternative is going down and down the spiral of democratic disenchantment and erosion…
The alternative is going down and down the spiral of democratic disenchantment and erosion…
We put cells and cytoskeleton filaments on the architecture of the musée d'Orsay.
www.musee-orsay.fr/fr/agenda/ev...
Scientists of the #CytoMorphoLab adapted their protocols to illustrate the questions that keep them awake at night.
-> Two shows on the 24th and 25th of January.


We put cells and cytoskeleton filaments on the architecture of the musée d'Orsay.
www.musee-orsay.fr/fr/agenda/ev...
Scientists of the #CytoMorphoLab adapted their protocols to illustrate the questions that keep them awake at night.
-> Two shows on the 24th and 25th of January.
Congratulations @manuelthery.bsky.social @lblanchoin.bsky.social and all Scientists of the #CytoMorphoLab for their poetic vision of the microscopic level.




Congratulations @manuelthery.bsky.social @lblanchoin.bsky.social and all Scientists of the #CytoMorphoLab for their poetic vision of the microscopic level.
The latest model of ChatGPT has begun to cite Elon Musk’s Grokipedia as a source on a wide range of queries, including on Iranian conglomerates and Holocaust deniers, raising concerns about misinformation on the platform.

www.nature.com/articles/s41...

www.nature.com/articles/s41...
We use them to map 665,856 pairwise genetic perturbations and outline a path to comprehensive interaction mapping in human cells.
We also introduce an approach for cloning lentiviral libraries with billions of elements.

We use them to map 665,856 pairwise genetic perturbations and outline a path to comprehensive interaction mapping in human cells.
We also introduce an approach for cloning lentiviral libraries with billions of elements.


