
Large-scale testing of antimicrobial lethality at single-cell resolution predicts mycobacterial infection outcomes.
From single cells to patients, across tuberculosis and M. abscessus.
👉 doi.org/10.1038/s415...
👇 Thread
Large-scale testing of antimicrobial lethality at single-cell resolution predicts mycobacterial infection outcomes.
From single cells to patients, across tuberculosis and M. abscessus.
👉 doi.org/10.1038/s415...
👇 Thread
Cambridge University, Department of Veterinary Medicine, 12/12/2025
"The decision to recommend the closure of what the Times Higher Education Supplement ranks as the best undergraduate veterinary course in the world has come as a bolt from the blue."
[1]
#SaveTheVetSchool
Cambridge University, Department of Veterinary Medicine, 12/12/2025
"The decision to recommend the closure of what the Times Higher Education Supplement ranks as the best undergraduate veterinary course in the world has come as a bolt from the blue."
[1]
#SaveTheVetSchool
Publication at NAR: doi.org/10.1093/nar/... @narjournal.bsky.social #microsky 🧵 1/8
Publication at NAR: doi.org/10.1093/nar/... @narjournal.bsky.social #microsky 🧵 1/8
We found that hospital patients were frequently colonised with P. aeruginosa and that the same clone was shared between the gut and the lung.
The phylogenies indicate that the clones moved from lung->gut
www.nature.com/articles/s41...
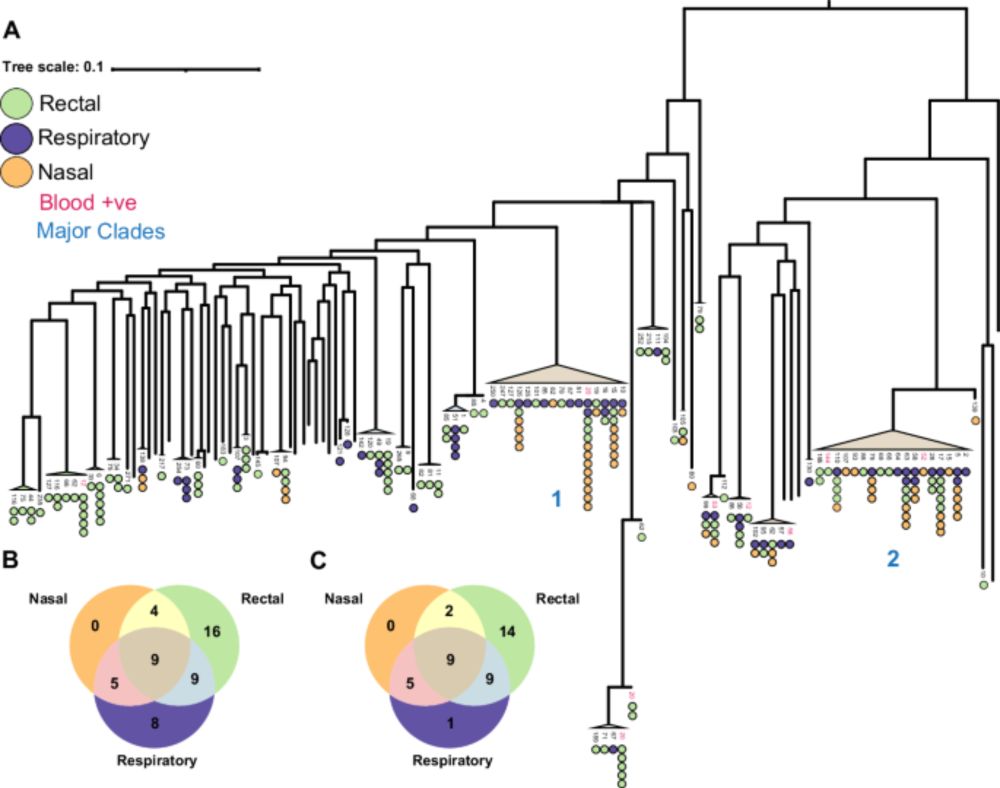
We found that hospital patients were frequently colonised with P. aeruginosa and that the same clone was shared between the gut and the lung.
The phylogenies indicate that the clones moved from lung->gut
www.nature.com/articles/s41...
These findings offer insight into who may be at higher risk of infection. 👇
www.sanger.ac.uk/news_item/la...

New study in @natmicrobiol.nature.com uncovers this hidden mechanism of drug resistance: buff.ly/s3YIwyK
@andresfloto.bsky.social @vpd-hlri.bsky.social @unibas.ch

New study in @natmicrobiol.nature.com uncovers this hidden mechanism of drug resistance: buff.ly/s3YIwyK
@andresfloto.bsky.social @vpd-hlri.bsky.social @unibas.ch
We're excited for users who might train new models, find phenotype/genotype mismatches, or any other use
EMBL-EBI’s new AMR portal brings together laboratory resistance data and bacterial genomes in one open platform.
#WAAW2025 #ActOnAMR
www.ebi.ac.uk/about/news/t...
🧬💻

We're excited for users who might train new models, find phenotype/genotype mismatches, or any other use
Review article published in @natrevmicro.nature.com with @bupbuse.bsky.social, Andriko von Kügelgen and @vikramalva.bsky.social.
S-layers are everywhere!
www.nature.com/articles/s41...

Review article published in @natrevmicro.nature.com with @bupbuse.bsky.social, Andriko von Kügelgen and @vikramalva.bsky.social.
S-layers are everywhere!
www.nature.com/articles/s41...
Metalog: curated and harmonised contextual data for global metagenomics samples
now out in @narjournal.bsky.social
academic.oup.com/nar/advance-...

Metalog: curated and harmonised contextual data for global metagenomics samples
now out in @narjournal.bsky.social
academic.oup.com/nar/advance-...
If you are at EMBL, ETH Zurich, GRC or GIMM working on a biological problem in data science or programming, consider putting a project forwards!

If you are at EMBL, ETH Zurich, GRC or GIMM working on a biological problem in data science or programming, consider putting a project forwards!
pseudomonas.io/home
pseudomonas.io/home
We’re looking for a PhD student to work at the interface of computational biophysics, machine learning & human mutations. 📌 FPI fellowship, 4 years fully funded!
More information here:
www.bsc.es/join-us/job-...
Thanks to all who have been part of this process!
tinyurl.com/4r9vf4zx

We’re looking for a PhD student to work at the interface of computational biophysics, machine learning & human mutations. 📌 FPI fellowship, 4 years fully funded!
More information here:
www.bsc.es/join-us/job-...
www.embl.org/about/info/e...
We have three positions in microbial genomics at EMBL-EBI, including one in my group. Please do apply, or if you know anyone that would be interested pass on to them
www.embl.org/about/info/e...
We have three positions in microbial genomics at EMBL-EBI, including one in my group. Please do apply, or if you know anyone that would be interested pass on to them
• 9× faster median speed vs bcftools
• Higher accuracy for both SNPs & indels
bioconda.github.io/recipes/quic...
bmcbiol.biomedcentral.com/articles/10....
#microsky #microbiomesky

• 9× faster median speed vs bcftools
• Higher accuracy for both SNPs & indels
bioconda.github.io/recipes/quic...
bmcbiol.biomedcentral.com/articles/10....
#microsky #microbiomesky
www.ebi.ac.uk/training/onl...
Including both molecular and epidemiological examples

www.ebi.ac.uk/training/onl...
Including both molecular and epidemiological examples
Combining a scalable human gut organoid model with TraDIS and statistical modeling, we mapped how Shigella colonizes the intestine at genome-wide resolution.
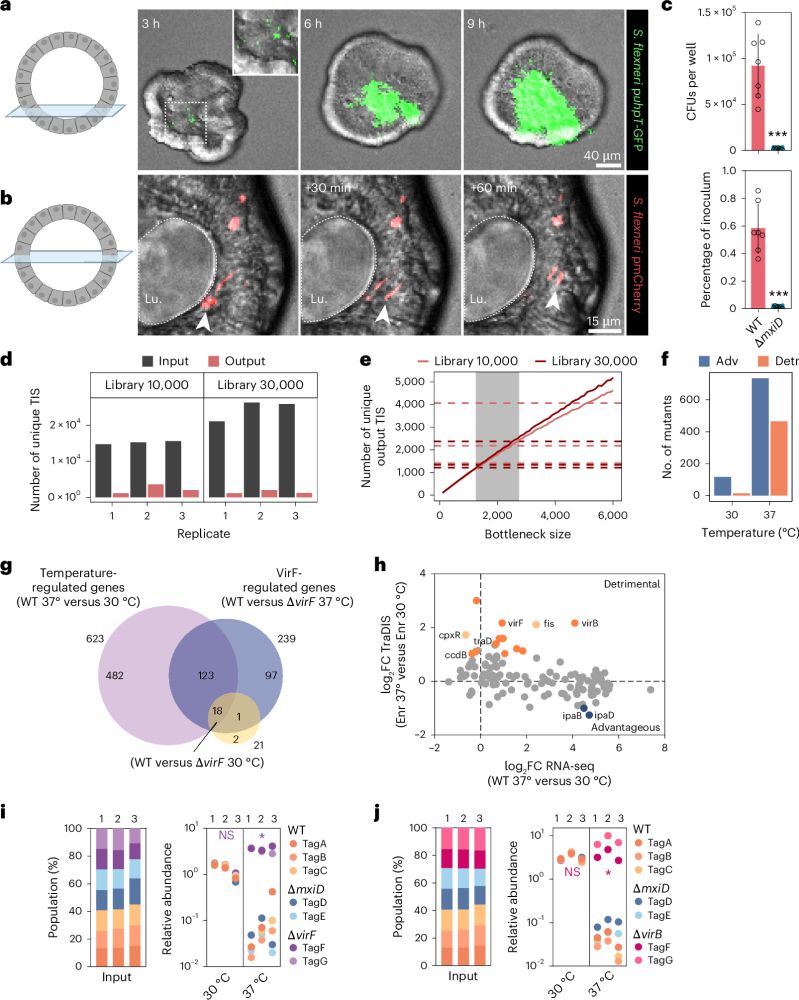
Combining a scalable human gut organoid model with TraDIS and statistical modeling, we mapped how Shigella colonizes the intestine at genome-wide resolution.
Preprint 📝: biorxiv.org/content/10.1...
🧵 1/n
Preprint 📝: biorxiv.org/content/10.1...
🧵 1/n
bsky.app/profile/macw...
Preprint 📝: biorxiv.org/content/10.1...
🧵 1/n

bsky.app/profile/macw...

More details below. 👇
More details below. 👇
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
www.cam.ac.uk/opinion-ai-a...

www.cam.ac.uk/opinion-ai-a...

