
go.rutgers.edu/ijwpxr1h

go.rutgers.edu/ijwpxr1h
Here are some #molstar renderings of an AgFe mixed metal Metal Organic Framework to celebrate.



Here are some #molstar renderings of an AgFe mixed metal Metal Organic Framework to celebrate.


Learn how to align one or more protein chains to a reference structure in a pairwise manner
pdb101.rcsb.org/news...

Learn how to align one or more protein chains to a reference structure in a pairwise manner
pdb101.rcsb.org/news...
ipymolstar repo: github.com/jhsmit/ipymo...
Molviewspec: molstar.org/mol-view-spec/
ipymolstar repo: github.com/jhsmit/ipymo...
Molviewspec: molstar.org/mol-view-spec/

The data is only encoded in the link and not stored so you can use it to share unpublished data*
(at your own risk, no warranty, links might stop working)

The data is only encoded in the link and not stored so you can use it to share unpublished data*
(at your own risk, no warranty, links might stop working)
Upload your .csv with residue/value data, select data column, colormap and norm to interactively color your protein model.
Then, add title/description and click the button to open a shareable view with url-encoded data and settings.
Upload your .csv with residue/value data, select data column, colormap and norm to interactively color your protein model.
Then, add title/description and click the button to open a shareable view with url-encoded data and settings.
#GeometryNodes #SciArt
#GeometryNodes #SciArt
www.khronos.org/events/webgl...
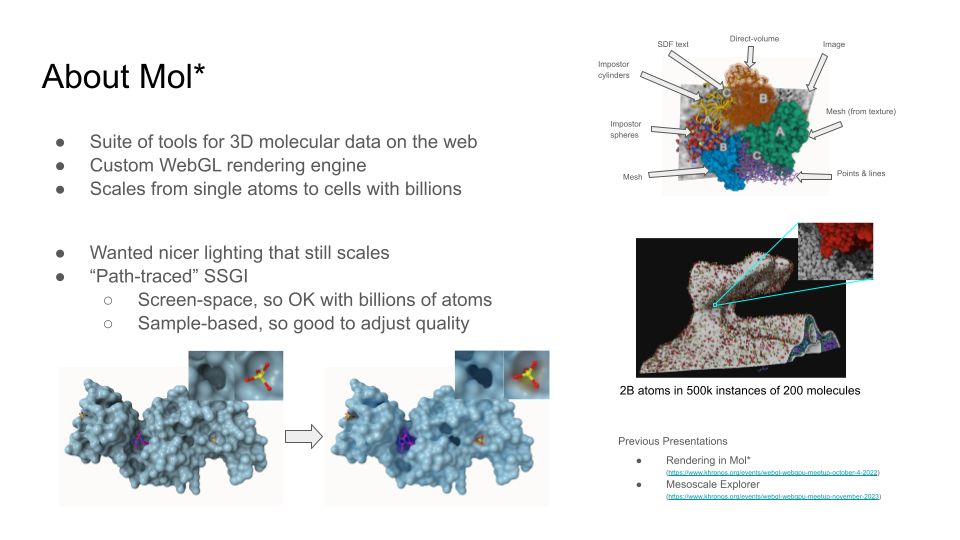
www.khronos.org/events/webgl...
AlphaBridge: A User-Friendly Tool for Interpreting Protein Predictions
Whether you're an expert or just new to the field of Protein Prediction, AlphaBridge is the way to go!
🔁Try it and share your thoughts!
📜 Article: www.biorxiv.org/content/10.1...
AlphaBridge: A User-Friendly Tool for Interpreting Protein Predictions
Whether you're an expert or just new to the field of Protein Prediction, AlphaBridge is the way to go!
🔁Try it and share your thoughts!
📜 Article: www.biorxiv.org/content/10.1...
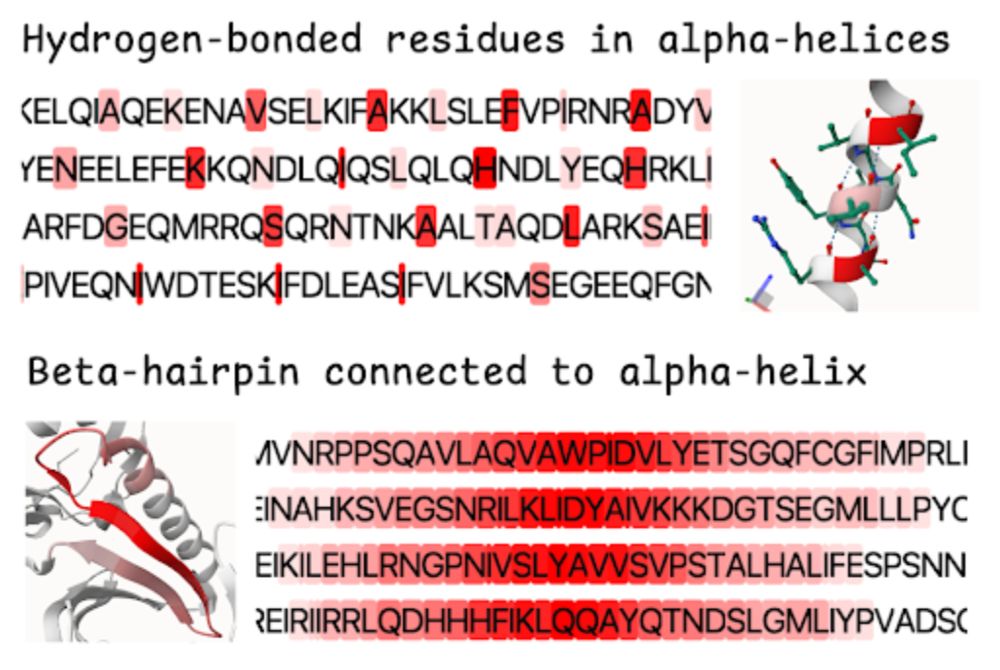
Try it on pycafe: py.cafe/jhsmit/ipymo...
Try it on pycafe: py.cafe/jhsmit/ipymo...


