
🇲🇽🇪🇺(🇦🇹🇫🇷🇪🇸) Symbionts, bacteria, insects, leeches, viruses, bioinformatics, evolution, genomics. Opinions are my very own.
#SymbioSky
https://besymblab.univie.ac.at/
https://ecoevo.social/@amanzanom

doi.org/10.12688/ope...
w/@ergabiodiv.bsky.social, @biogeneurope.bsky.social, @leibnizlib.bsky.social, & @sangerinstitute.bsky.social
@dome-vienna.bsky.social, @cemess.bsky.social, @univie.ac.at
1/3..
A secreted #endosymbiont protein (SyeA, formally Yba3) essential for colonizing host cells
doi.org/10.1101/2025...
#SymbioSky

A secreted #endosymbiont protein (SyeA, formally Yba3) essential for colonizing host cells
doi.org/10.1101/2025...
#SymbioSky
We’ve just released GLADE, a tool to infer gene gains, losses, duplications, and ancestral genomes across a phylogeny.
GLADE runs directly on OrthoFinder results.
www.biorxiv.org/content/10.6...
github.com/lauriebelch/...
(1/10)
We’ve just released GLADE, a tool to infer gene gains, losses, duplications, and ancestral genomes across a phylogeny.
GLADE runs directly on OrthoFinder results.
www.biorxiv.org/content/10.6...
github.com/lauriebelch/...
(1/10)

academic.oup.com/bioinformati...

academic.oup.com/bioinformati...
doi.org/10.12688/ope...
w/@ergabiodiv.bsky.social, @biogeneurope.bsky.social, @leibnizlib.bsky.social, & @sangerinstitute.bsky.social
@dome-vienna.bsky.social, @cemess.bsky.social, @univie.ac.at
1/3..

doi.org/10.12688/ope...
w/@ergabiodiv.bsky.social, @biogeneurope.bsky.social, @leibnizlib.bsky.social, & @sangerinstitute.bsky.social
@dome-vienna.bsky.social, @cemess.bsky.social, @univie.ac.at
1/3..
#TransparentPeerReview
Co-evolving infectivity and expression patterns drive the diversification of endogenous retroviruses
@juliusbrennecke.bsky.social et al
www.embopress.org/doi/full/10....
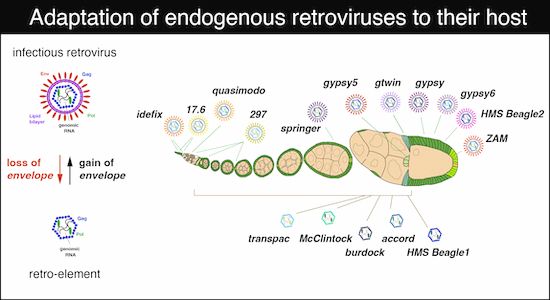
🔗 doi.org/10.1093/molbev/msaf238
#evobio #molbio

🔗 doi.org/10.1093/molbev/msaf238
#evobio #molbio
#VirEvol

#VirEvol
Alongside the new work on gutless worms, our study shows that organosulfur cycling is also essential in lucinid holobionts. Lucinid host provides DMSP to its symbionts, sulphur-oxidising Thiodiazotropha and, a new member, Endozoicomonas.

Alongside the new work on gutless worms, our study shows that organosulfur cycling is also essential in lucinid holobionts. Lucinid host provides DMSP to its symbionts, sulphur-oxidising Thiodiazotropha and, a new member, Endozoicomonas.
The hard work of @pegah-kalatehjari.bsky.social already paying off. Soon to come, baby leeches @dome-vienna.bsky.social
@fwf-at.bsky.social @univie.ac.at

The hard work of @pegah-kalatehjari.bsky.social already paying off. Soon to come, baby leeches @dome-vienna.bsky.social
@fwf-at.bsky.social @univie.ac.at
Natural and artificial variations of the standard #geneticCode
Of course with mention of #endosymbiont lineages
doi.org/10.1016/j.cu...

Natural and artificial variations of the standard #geneticCode
Of course with mention of #endosymbiont lineages
doi.org/10.1016/j.cu...
Have a read!! ➡️ doi.org/10.1093/gbe/...
@emilruff.bsky.social @alexjaffe.bsky.social @geomicrosoares.bsky.social & others not in Bluesky!

Have a read!! ➡️ doi.org/10.1093/gbe/...
@emilruff.bsky.social @alexjaffe.bsky.social @geomicrosoares.bsky.social & others not in Bluesky!
a 🧵 1/n
Drain: arxiv.org/abs/2511.04820
Strain: direct.mit.edu/qss/article/...
Oligopoly: direct.mit.edu/qss/article/...




a 🧵 1/n
Drain: arxiv.org/abs/2511.04820
Strain: direct.mit.edu/qss/article/...
Oligopoly: direct.mit.edu/qss/article/...

vConTACT3 delivers a unified, scalable, and transparent framework for genome-based virus taxonomy — helping translate big viral data into systematic classification.
🔗 Read the preprint: doi.org/10.1101/2025...
Improvements details below 👇

vConTACT3 delivers a unified, scalable, and transparent framework for genome-based virus taxonomy — helping translate big viral data into systematic classification.
🔗 Read the preprint: doi.org/10.1101/2025...
Improvements details below 👇
2) Parallel #evolution of #Bacteroidota as Long-Term #endosymbionts of #insects
by Jinyeong Choi, Cong Liu, Pradeep Palanichamy, Yumiko Masukagami, Hirotaka Tanaka, Takumasa Kondo, Matthew E. Gruwell, @filiphusnik.bsky.social
doi.org/10.1101/2025...
#SymbioSky @oistedu.bsky.social

2) Parallel #evolution of #Bacteroidota as Long-Term #endosymbionts of #insects
by Jinyeong Choi, Cong Liu, Pradeep Palanichamy, Yumiko Masukagami, Hirotaka Tanaka, Takumasa Kondo, Matthew E. Gruwell, @filiphusnik.bsky.social
doi.org/10.1101/2025...
#SymbioSky @oistedu.bsky.social
1) Genomic diversity and functional potential of facultative bacterial #symbionts across #scaleInsects
by Pradeep Palanichamy, Jinyeong Choi, Arno Hagenbeek
doi.org/10.1101/2025...
#SymbioSky

1) Genomic diversity and functional potential of facultative bacterial #symbionts across #scaleInsects
by Pradeep Palanichamy, Jinyeong Choi, Arno Hagenbeek
doi.org/10.1101/2025...
#SymbioSky
Defensive fungal symbiosis on insect hindlegs: www.science.org/doi/10.1126/...
#SymbioSky

Defensive fungal symbiosis on insect hindlegs: www.science.org/doi/10.1126/...
#SymbioSky
Come work with us to investigate how innate immunity works in an important agricultural pest, and disentangle how the immune system evolves. 🇬🇧🇨🇳
For more info, see: bit.ly/4ntrsEe
#Drosophila #Aphid #Immunity

Come work with us to investigate how innate immunity works in an important agricultural pest, and disentangle how the immune system evolves. 🇬🇧🇨🇳
For more info, see: bit.ly/4ntrsEe
#Drosophila #Aphid #Immunity
bmcgenomics.biomedcentral.com/articles/10....

bmcgenomics.biomedcentral.com/articles/10....
#SymbioSky
doi.org/10.1111/1758...

#SymbioSky
doi.org/10.1111/1758...
~27,000 predicted viral protein monomers & homodimers
Conserved folds across bacteria, archaea & eukaryotic viruses
New toxin–antitoxin system KreTA uncovered
Vast “functional darkness” remains uncharted
www.science.org/doi/10.1126/...

~27,000 predicted viral protein monomers & homodimers
Conserved folds across bacteria, archaea & eukaryotic viruses
New toxin–antitoxin system KreTA uncovered
Vast “functional darkness” remains uncharted
www.science.org/doi/10.1126/...
Latest for @nytimes.com on a fantastic #fossil: www.nytimes.com/2025/10/01/s...

Latest for @nytimes.com on a fantastic #fossil: www.nytimes.com/2025/10/01/s...

