Abraham Gihawi
@abrahamgihawi.bsky.social
UEA Research Fellow - Bioinformatics - Cancer (meta)genomics since 2017.
Opinions are mine alone.
Opinions are mine alone.
Reposted by Abraham Gihawi
🚀 TaxSEA v1.2 is now live in #Bioconductor release 3.22!
TaxSEA brings enrichment testing to #metagenomics data. Just like GSEA does for transcriptomics making #microbiome & #microbiota results biologically interpretable, fast, and reproducible. We've added new DBs & features in 1.2 👇 🧵
TaxSEA brings enrichment testing to #metagenomics data. Just like GSEA does for transcriptomics making #microbiome & #microbiota results biologically interpretable, fast, and reproducible. We've added new DBs & features in 1.2 👇 🧵
November 3, 2025 at 12:27 AM
🚀 TaxSEA v1.2 is now live in #Bioconductor release 3.22!
TaxSEA brings enrichment testing to #metagenomics data. Just like GSEA does for transcriptomics making #microbiome & #microbiota results biologically interpretable, fast, and reproducible. We've added new DBs & features in 1.2 👇 🧵
TaxSEA brings enrichment testing to #metagenomics data. Just like GSEA does for transcriptomics making #microbiome & #microbiota results biologically interpretable, fast, and reproducible. We've added new DBs & features in 1.2 👇 🧵
Reposted by Abraham Gihawi
Dear community, Bakta needs your help!
To further improve the functional annotation of "hypothetical" CDS, me and @gbouras13.bsky.social, we are looking for the worst Bakta-annotated bacterial genomes ;-)
(1/2)
To further improve the functional annotation of "hypothetical" CDS, me and @gbouras13.bsky.social, we are looking for the worst Bakta-annotated bacterial genomes ;-)
(1/2)
October 6, 2025 at 7:27 AM
Dear community, Bakta needs your help!
To further improve the functional annotation of "hypothetical" CDS, me and @gbouras13.bsky.social, we are looking for the worst Bakta-annotated bacterial genomes ;-)
(1/2)
To further improve the functional annotation of "hypothetical" CDS, me and @gbouras13.bsky.social, we are looking for the worst Bakta-annotated bacterial genomes ;-)
(1/2)
Reposted by Abraham Gihawi
📣 Speakers announced for the Oral Tissue Collaborative (OTC) Symposium
Register now to join either in person or online for the event on the future of oral tissue biorepositories
🗓️ 10 October
Register by 3 October ⤵️
Register now to join either in person or online for the event on the future of oral tissue biorepositories
🗓️ 10 October
Register by 3 October ⤵️

Oral Tissue Collaborative (OTC) Symposium: Building Excellence in Oral Tissue Biobanking - Quadram Institute
This symposium brings together leading experts, researchers, and practitioners in the field of oral tissue biobanking
buff.ly
September 25, 2025 at 8:01 AM
📣 Speakers announced for the Oral Tissue Collaborative (OTC) Symposium
Register now to join either in person or online for the event on the future of oral tissue biorepositories
🗓️ 10 October
Register by 3 October ⤵️
Register now to join either in person or online for the event on the future of oral tissue biorepositories
🗓️ 10 October
Register by 3 October ⤵️
Reposted by Abraham Gihawi
Delighted to finally announce a preprint describing the Q100 project! “A complete diploid human genome benchmark for personalized genomics” For which we finished HG002 to near-perfect accuracy: www.biorxiv.org/content/10.1... 🧵[1/14]

A complete diploid human genome benchmark for personalized genomics
Human genome resequencing typically involves mapping reads to a reference genome to call variants; however, this approach suffers from both technical and reference biases, leaving many duplicated and ...
www.biorxiv.org
September 22, 2025 at 5:01 PM
Delighted to finally announce a preprint describing the Q100 project! “A complete diploid human genome benchmark for personalized genomics” For which we finished HG002 to near-perfect accuracy: www.biorxiv.org/content/10.1... 🧵[1/14]
Reposted by Abraham Gihawi
Spacedust: a tool for de novo identification of conserved gene clusters from metagenomic data.
@ruoshiz.bsky.social @milot.bsky.social
www.nature.com/articles/s41...
@ruoshiz.bsky.social @milot.bsky.social
www.nature.com/articles/s41...

De novo discovery of conserved gene clusters in microbial genomes with Spacedust - Nature Methods
This work presents Spacedust, a tool for de novo identification of conserved gene clusters from metagenomic data.
www.nature.com
September 16, 2025 at 1:25 PM
Spacedust: a tool for de novo identification of conserved gene clusters from metagenomic data.
@ruoshiz.bsky.social @milot.bsky.social
www.nature.com/articles/s41...
@ruoshiz.bsky.social @milot.bsky.social
www.nature.com/articles/s41...
Finding microbes in cancer sequencing data has been controversial. Are we seeing real biology… or just contamination + batch effects?
September 3, 2025 at 7:00 PM
Finding microbes in cancer sequencing data has been controversial. Are we seeing real biology… or just contamination + batch effects?
Reposted by Abraham Gihawi
Two papers in today's issue of @nature.com : 1) we assemble 65 genomes to near completion, including centromeres and the MHC. tinyurl.com/3huhax6w. 2) we sequence 1,019 genomes from the 1kGP with long reads, revealing SVs down to low allele frequencies tinyurl.com/wbx3we9x.
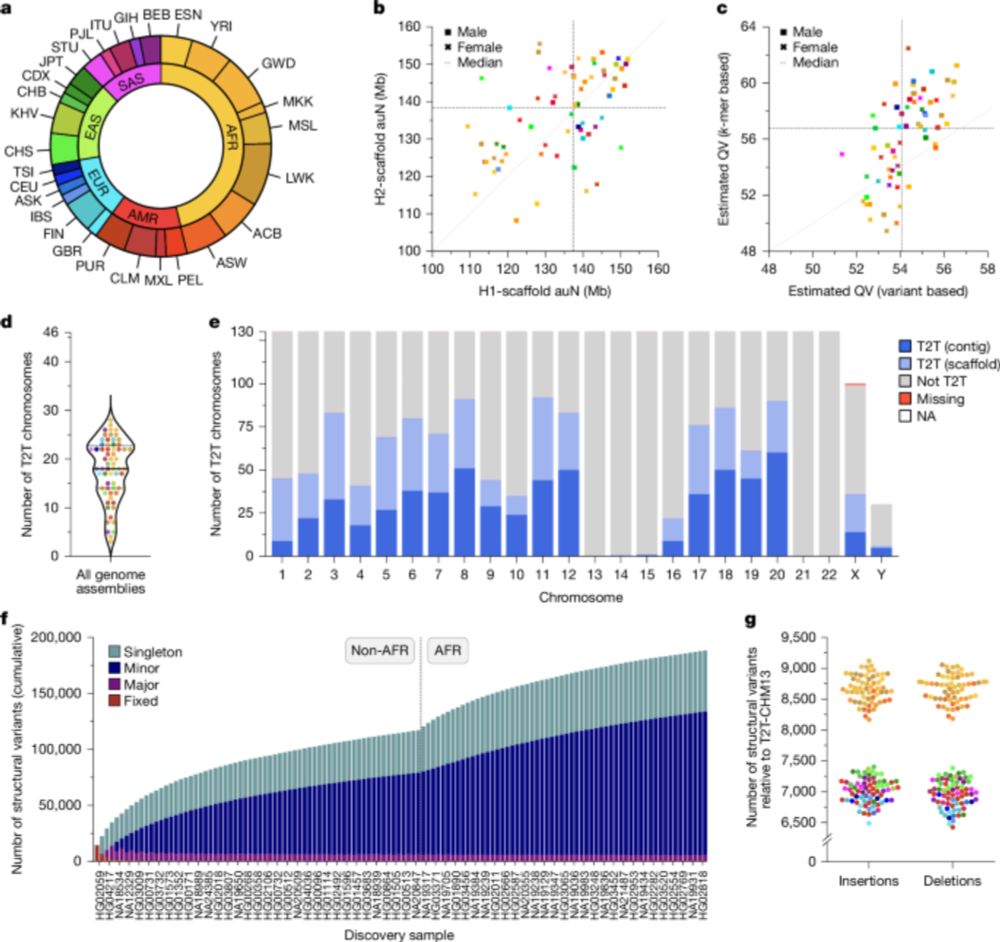
Complex genetic variation in nearly complete human genomes - Nature
Using sequencing and haplotype-resolved assembly of 65 diverse human genomes, complex regions including the major histocompatibility complex and centromeres are analysed.
tinyurl.com
July 23, 2025 at 3:12 PM
Two papers in today's issue of @nature.com : 1) we assemble 65 genomes to near completion, including centromeres and the MHC. tinyurl.com/3huhax6w. 2) we sequence 1,019 genomes from the 1kGP with long reads, revealing SVs down to low allele frequencies tinyurl.com/wbx3we9x.
Reposted by Abraham Gihawi
About 18 months ago we held a @microbiologysociety.org meeting in Glasgow to discuss the thorny topic of #microbial #taxonomy and #nomenclature - and now you can read what we talked about :D
#microbiology #controversy
doi.org/10.1099/ijse...
#microbiology #controversy
doi.org/10.1099/ijse...

‘What’s in a name? Fit-for-purpose bacterial nomenclature’: meeting report
Rapid and economical DNA sequencing has resulted in a revolution in phylogenomics. The impact of changes in nomenclature can be perceived as an absolute necessity of scientific rigour, coupled with th...
doi.org
July 23, 2025 at 4:51 PM
About 18 months ago we held a @microbiologysociety.org meeting in Glasgow to discuss the thorny topic of #microbial #taxonomy and #nomenclature - and now you can read what we talked about :D
#microbiology #controversy
doi.org/10.1099/ijse...
#microbiology #controversy
doi.org/10.1099/ijse...
Reposted by Abraham Gihawi
Hope this is useful - consensus statement "Guidelines for preventing and reporting contamination in low-biomass microbiome studies" rdcu.be/er3Io
Guidelines for preventing and reporting contamination in low-biomass microbiome studies
Nature Microbiology - In this Consensus Statement, the authors outline strategies for processing, analysing and interpreting low-biomass microbiome samples, and provide recommendations to minimize...
rdcu.be
June 21, 2025 at 7:59 PM
Hope this is useful - consensus statement "Guidelines for preventing and reporting contamination in low-biomass microbiome studies" rdcu.be/er3Io
Reposted by Abraham Gihawi
I'm happy to announce the latest release of the GlobDB, available at globdb.org.
The GlobDB is a database of "species dereplicated" microbial genomes, and as of release 226 contains twice the number of species-representative genomes (306,260) than the latest GTDB release.
The GlobDB is a database of "species dereplicated" microbial genomes, and as of release 226 contains twice the number of species-representative genomes (306,260) than the latest GTDB release.
home | GlobDB
globdb.org
June 10, 2025 at 11:20 AM
I'm happy to announce the latest release of the GlobDB, available at globdb.org.
The GlobDB is a database of "species dereplicated" microbial genomes, and as of release 226 contains twice the number of species-representative genomes (306,260) than the latest GTDB release.
The GlobDB is a database of "species dereplicated" microbial genomes, and as of release 226 contains twice the number of species-representative genomes (306,260) than the latest GTDB release.
Reposted by Abraham Gihawi
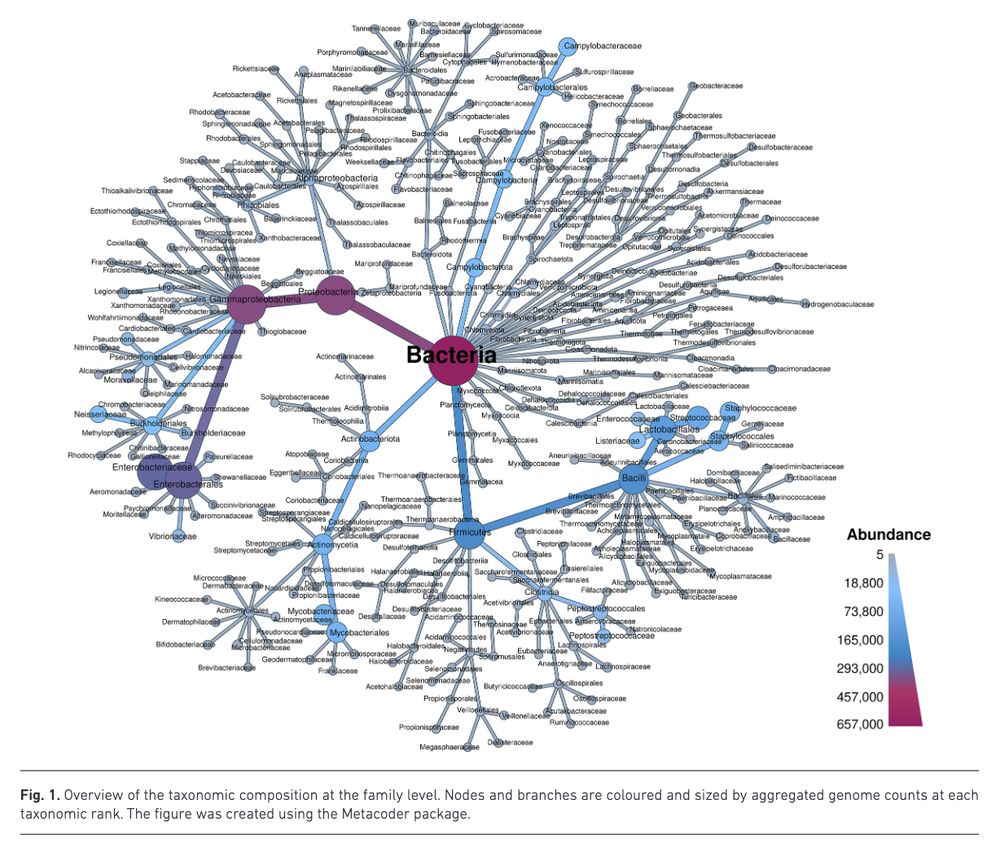
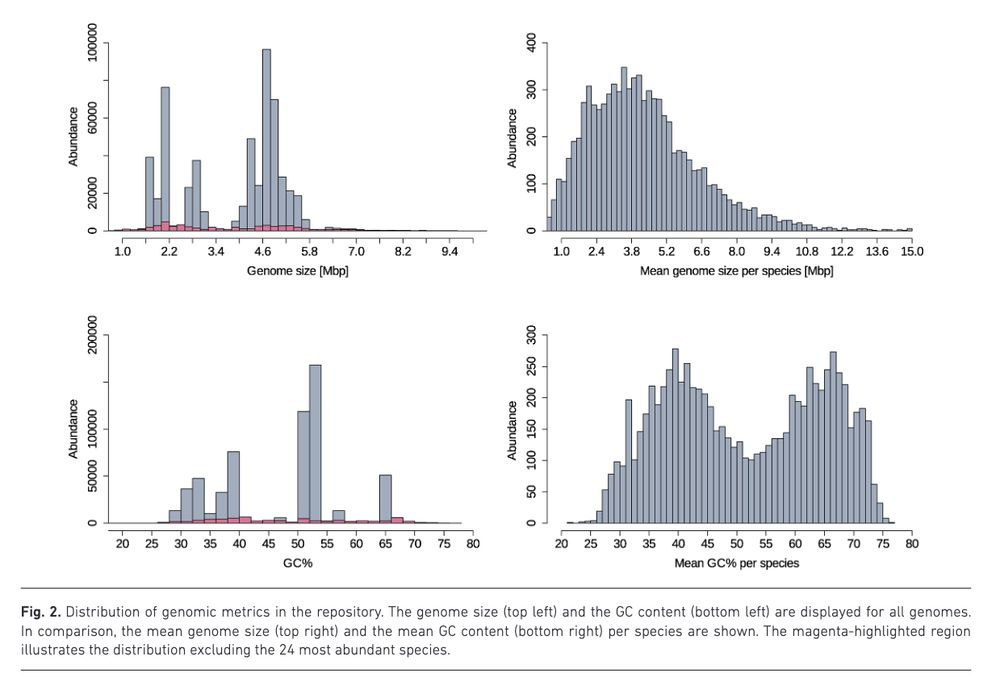
BakRep – a searchable large-scale web repository for bacterial genomes, characterizations and metadata www.microbiologyresearch.org/content/jour... 🧬🖥️ Nextflow code: github.com/ag-computati...
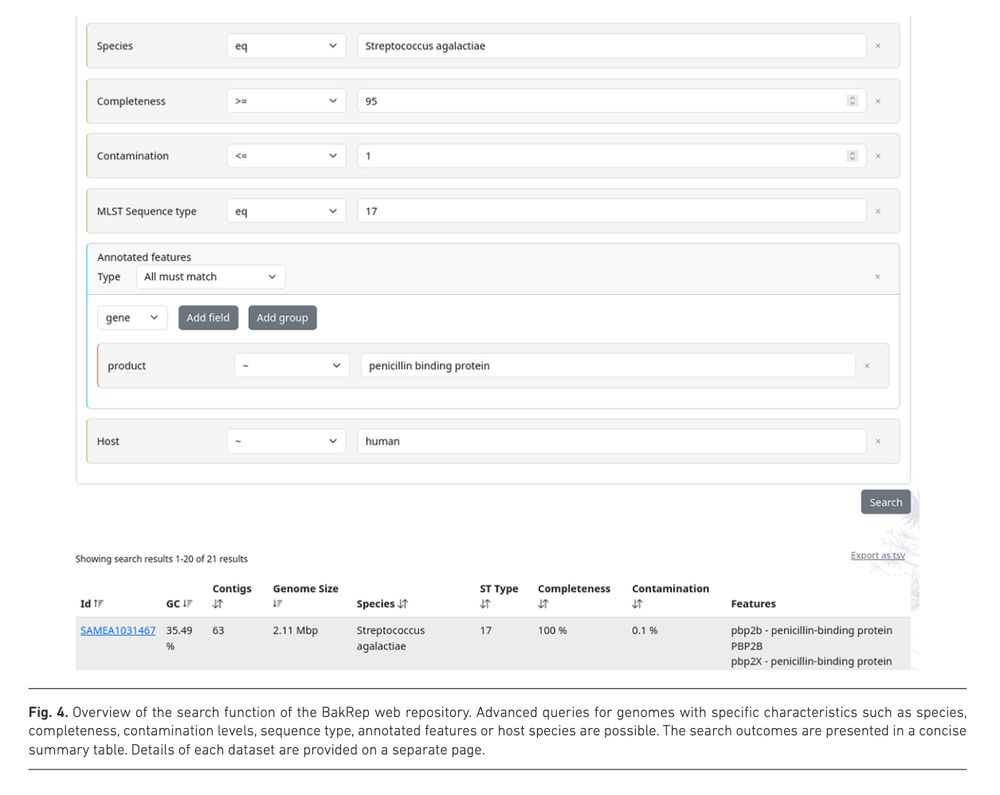
June 4, 2025 at 8:03 PM
BakRep – a searchable large-scale web repository for bacterial genomes, characterizations and metadata www.microbiologyresearch.org/content/jour... 🧬🖥️ Nextflow code: github.com/ag-computati...
Reposted by Abraham Gihawi
New preprint! Autocycler is a tool for long-read consensus assembly of bacterial genomes. It's like Trycycler but can be run fully automated (without any human intervention).
www.biorxiv.org/content/10.1...
(1/6)
www.biorxiv.org/content/10.1...
(1/6)

Autocycler: long-read consensus assembly for bacterial genomes
Motivation Long-read sequencing enables complete bacterial genome assemblies, but individual assemblers are imperfect and often produce sequence-level and structural errors. Consensus assembly using T...
www.biorxiv.org
May 21, 2025 at 3:29 AM
New preprint! Autocycler is a tool for long-read consensus assembly of bacterial genomes. It's like Trycycler but can be run fully automated (without any human intervention).
www.biorxiv.org/content/10.1...
(1/6)
www.biorxiv.org/content/10.1...
(1/6)
Reposted by Abraham Gihawi
The human pangenome continues to grow and improve! Release 2 is here! Click through for the details, but this is a pretty amazing dataset including not just the phased assemblies, but PacBio HiFi, ONT Ultralong, Dovetail/Illumina Hi-C, PacBio Kinnex, and Illumina WGS for all samples
📢 HPRC Release 2 is here!
Now with phased genomes from 200+ individuals, a 5x increase from Release 1.
Explore sequencing data, assemblies, annotations & alignments in our interactive data explorer ⬇️:
humanpangenome.org/hprc-data-re...
Now with phased genomes from 200+ individuals, a 5x increase from Release 1.
Explore sequencing data, assemblies, annotations & alignments in our interactive data explorer ⬇️:
humanpangenome.org/hprc-data-re...

May 12, 2025 at 1:52 PM
The human pangenome continues to grow and improve! Release 2 is here! Click through for the details, but this is a pretty amazing dataset including not just the phased assemblies, but PacBio HiFi, ONT Ultralong, Dovetail/Illumina Hi-C, PacBio Kinnex, and Illumina WGS for all samples
Reposted by Abraham Gihawi
This is important work for the exciting next stage of metagenomics #MicroSky
I am very happy (and anxious) to share with you our most recent work in which we evaluated four of the most popular long-read assemblers,
www.biorxiv.org/content/10.1...
and tell you just a little bit about it in the following 🧵
www.biorxiv.org/content/10.1...
and tell you just a little bit about it in the following 🧵

Assemblies of long-read metagenomes suffer from diverse errors
Genomes from metagenomes have revolutionised our understanding of microbial diversity, ecology, and evolution, propelling advances in basic science, biomedicine, and biotechnology. Assembly algorithms...
www.biorxiv.org
April 28, 2025 at 11:04 AM
This is important work for the exciting next stage of metagenomics #MicroSky
Reposted by Abraham Gihawi
Very compelling study showing the association between herpes zoster vaccination and dementia prevention. The authors leveraged a unique cohort for this natural experiment which shows a reduction in dementia risk of 20% in vaccinated individuals. #MedSky #IDSky 🧪
www.nature.com/articles/s41...
www.nature.com/articles/s41...
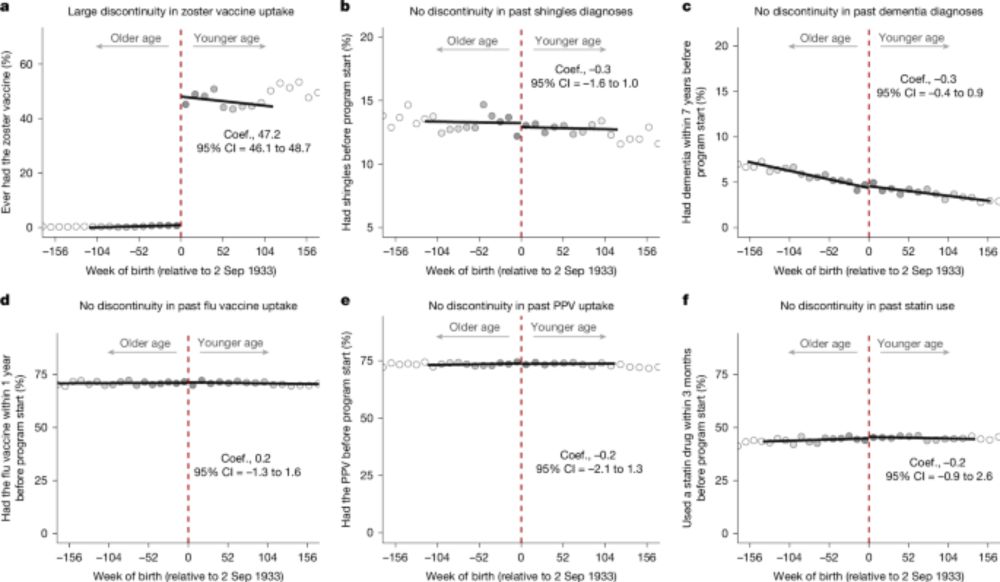
A natural experiment on the effect of herpes zoster vaccination on dementia - Nature
Using a natural experiment that avoids common bias concerns, this study finds that the live-attenuated shingles vaccine reduced the probability of a new dementia diagnosis within a follow-up period of...
www.nature.com
April 2, 2025 at 5:49 PM
Very compelling study showing the association between herpes zoster vaccination and dementia prevention. The authors leveraged a unique cohort for this natural experiment which shows a reduction in dementia risk of 20% in vaccinated individuals. #MedSky #IDSky 🧪
www.nature.com/articles/s41...
www.nature.com/articles/s41...
Reposted by Abraham Gihawi
If you generate, or use MAGs, you should really read the work we have done benchmarking their limitations. Using complex mock communities (70 species), we assessed how deeply you need to sequence to accurately conduct common metaG analyses. Webwere shocked to see even HQ MAGs are often chimeric!
Benchmarking of shotgun sequencing depth highlights strain-level limitations of metagenomic analysis https://www.biorxiv.org/content/10.1101/2025.03.27.645659v1
March 28, 2025 at 9:17 AM
If you generate, or use MAGs, you should really read the work we have done benchmarking their limitations. Using complex mock communities (70 species), we assessed how deeply you need to sequence to accurately conduct common metaG analyses. Webwere shocked to see even HQ MAGs are often chimeric!
Reposted by Abraham Gihawi
🧫🧪 "Ten species comprise half of the bacteriology literature,
leaving most species unstudied."
'Model' gram - and gram + (Escherichia coli and Bacillus subtitles) + human pathogens
www.biorxiv.org/content/10.1...
leaving most species unstudied."
'Model' gram - and gram + (Escherichia coli and Bacillus subtitles) + human pathogens
www.biorxiv.org/content/10.1...

March 6, 2025 at 5:03 PM
🧫🧪 "Ten species comprise half of the bacteriology literature,
leaving most species unstudied."
'Model' gram - and gram + (Escherichia coli and Bacillus subtitles) + human pathogens
www.biorxiv.org/content/10.1...
leaving most species unstudied."
'Model' gram - and gram + (Escherichia coli and Bacillus subtitles) + human pathogens
www.biorxiv.org/content/10.1...
Reposted by Abraham Gihawi
1/9 🚨 New preprint alert!
Frustrated by strange results with your metagenomic data?
Meet CroCoDeEL, an accurate, easy-to-use tool to detect and quantify cross-sample contamination—no need for negative controls or sample positions!
🔗 doi.org/10.1101/2025...
Frustrated by strange results with your metagenomic data?
Meet CroCoDeEL, an accurate, easy-to-use tool to detect and quantify cross-sample contamination—no need for negative controls or sample positions!
🔗 doi.org/10.1101/2025...

CroCoDeEL: accurate control-free detection of cross-sample contamination in metagenomic data
Metagenomic sequencing provides profound insights into microbial communities, but it is often compromised by technical biases, including cross-sample contamination. This underexplored phenomenon arise...
doi.org
January 20, 2025 at 12:42 PM
1/9 🚨 New preprint alert!
Frustrated by strange results with your metagenomic data?
Meet CroCoDeEL, an accurate, easy-to-use tool to detect and quantify cross-sample contamination—no need for negative controls or sample positions!
🔗 doi.org/10.1101/2025...
Frustrated by strange results with your metagenomic data?
Meet CroCoDeEL, an accurate, easy-to-use tool to detect and quantify cross-sample contamination—no need for negative controls or sample positions!
🔗 doi.org/10.1101/2025...
Reposted by Abraham Gihawi
Today we report that an engineered skin bacterium, swabbed gently on the head of a mouse, can unleash a potent antibody response against a pathogen. Could lead to topical vaccines that are applied in a cream. @djenetbousbaine.bsky.social led the charge... @natureportfolio.bsky.social 1/55

December 11, 2024 at 4:29 PM
Today we report that an engineered skin bacterium, swabbed gently on the head of a mouse, can unleash a potent antibody response against a pathogen. Could lead to topical vaccines that are applied in a cream. @djenetbousbaine.bsky.social led the charge... @natureportfolio.bsky.social 1/55
Reposted by Abraham Gihawi
This seems odd. A cancer microbiome machine learning paper that uses data from the retracted Poore et al. 2020 Nature paper has been published just last week in mSystems. Some of the claims seem very strong. Also, it seems to have been accepted four days after submission.
🧬 & 🖥️ #microbiome
🧬 & 🖥️ #microbiome

December 2, 2024 at 2:54 PM
This seems odd. A cancer microbiome machine learning paper that uses data from the retracted Poore et al. 2020 Nature paper has been published just last week in mSystems. Some of the claims seem very strong. Also, it seems to have been accepted four days after submission.
🧬 & 🖥️ #microbiome
🧬 & 🖥️ #microbiome
Reposted by Abraham Gihawi
This has been a long journey and I think it's the correct outcome. Thanks to everyone that has been involved, especially @abrahamgihawi.bsky.social who has been investigating this since his PhD days, and Steven Salzberg for really raising the analysis and awareness to another level.
🧬🖥️
🧬🖥️

June 27, 2024 at 12:47 PM
This has been a long journey and I think it's the correct outcome. Thanks to everyone that has been involved, especially @abrahamgihawi.bsky.social who has been investigating this since his PhD days, and Steven Salzberg for really raising the analysis and awareness to another level.
🧬🖥️
🧬🖥️
Reposted by Abraham Gihawi
Great PhD opportunity available with us. Bacteria, bioinformatics, lab work, ancestry, and prostate cancer. Fully funded for those eligible for UK fees. Deadline 15th April.
buff.ly/43AZD4k
@ueamedical.bsky.social
🧬🖥️🩺 #microbiome
Contact me or @abrahamgihawi.bsky.social for more details.
buff.ly/43AZD4k
@ueamedical.bsky.social
🧬🖥️🩺 #microbiome
Contact me or @abrahamgihawi.bsky.social for more details.

Aggressive prostate cancer in African men: are microbes involved? Investigating decontamination, microbiomes and human genomics (BREWER_U24MMB) at University of East Anglia on FindAPhD.com
PhD Project - Aggressive prostate cancer in African men: are microbes involved? Investigating decontamination, microbiomes and human genomics (BREWER_U24MMB) at University of East Anglia, listed on Fi...
buff.ly
March 28, 2024 at 9:22 AM
Great PhD opportunity available with us. Bacteria, bioinformatics, lab work, ancestry, and prostate cancer. Fully funded for those eligible for UK fees. Deadline 15th April.
buff.ly/43AZD4k
@ueamedical.bsky.social
🧬🖥️🩺 #microbiome
Contact me or @abrahamgihawi.bsky.social for more details.
buff.ly/43AZD4k
@ueamedical.bsky.social
🧬🖥️🩺 #microbiome
Contact me or @abrahamgihawi.bsky.social for more details.
Reposted by Abraham Gihawi
New review paper out today in Journal of Medical Biology "Cancer invasion and anaerobic bacteria: new insights into mechanisms". Great work by Rachel Hurst, with @abrahamgihawi.bsky.social, John Wain, and Colin Cooper.
doi.org/10.1099/jmm....
doi.org/10.1099/jmm....

March 27, 2024 at 3:35 PM
New review paper out today in Journal of Medical Biology "Cancer invasion and anaerobic bacteria: new insights into mechanisms". Great work by Rachel Hurst, with @abrahamgihawi.bsky.social, John Wain, and Colin Cooper.
doi.org/10.1099/jmm....
doi.org/10.1099/jmm....
Reposted by Abraham Gihawi
New paper by Pat Schloss about rarefaction is definitely worth a read for microbiome aficionados
journals.asm.org/doi/10.1128/...
journals.asm.org/doi/10.1128/...
Waste not, want not: revisiting the analysis that called into question the practice of rarefaction |...
Over the past 10 years, the best method for normalizing the sequencing depth of samples
characterized by 16S rRNA gene sequencing has been contentious. An often cited article
by McMurdie and Holmes fo...
journals.asm.org
December 7, 2023 at 1:51 PM
New paper by Pat Schloss about rarefaction is definitely worth a read for microbiome aficionados
journals.asm.org/doi/10.1128/...
journals.asm.org/doi/10.1128/...
Come and join our group as a PhD student using computational methods to investigate bacteria and viruses in aggressive prostate cancer. Applications are now open! @danbrewer.bsky.social www.findaphd.com/phds/project...
October 30, 2023 at 3:55 PM
Come and join our group as a PhD student using computational methods to investigate bacteria and viruses in aggressive prostate cancer. Applications are now open! @danbrewer.bsky.social www.findaphd.com/phds/project...

