
she/her | IPA: ˈafkə ɡrɔs

@zhaoshh.bsky.social and I ask the question that Euler left open: How does an elastic line buckle within a (general) curved surface?
@mpipks.bsky.social @mpi-cbg.de @csbdresden.bsky.social
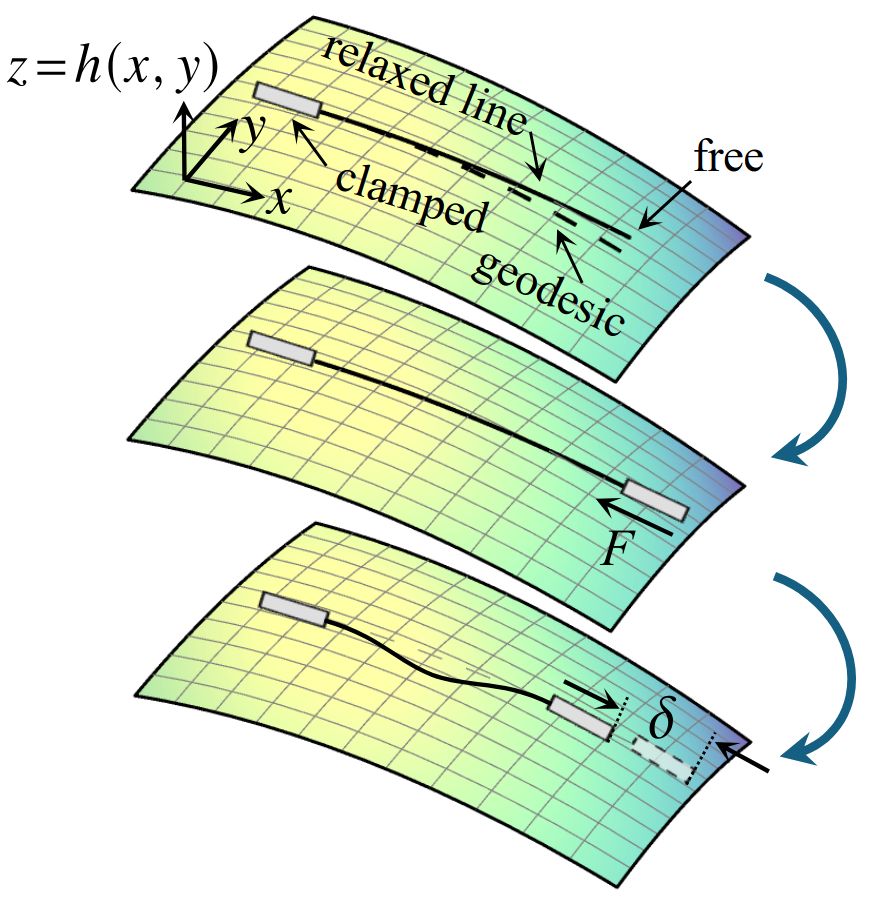
@zhaoshh.bsky.social and I ask the question that Euler left open: How does an elastic line buckle within a (general) curved surface?
@mpipks.bsky.social @mpi-cbg.de @csbdresden.bsky.social
This release focuses on improvements in clusterization and simplification as well as stabilization. Here's a release announcement with more details on past, present and future; please RT!
meshoptimizer.org/v1
This release focuses on improvements in clusterization and simplification as well as stabilization. Here's a release announcement with more details on past, present and future; please RT!
meshoptimizer.org/v1
It was hard not to load our building and around it into #bigvolumebrowser (22 M points + Fire LUT for height)
It was hard not to load our building and around it into #bigvolumebrowser (22 M points + Fire LUT for height)
forum.image.sc/t/bigvolumeb...

forum.image.sc/t/bigvolumeb...
It is the first deep-learning-based ("AI") drop-in replacement for TrackMate's regular LAP-Track algorithm.
Tracks out of the box for many types of datasets (bacteria, fluorescent nuclei, phase contrast cell culture etc), zero parameters to tune 🤖
It is the first deep-learning-based ("AI") drop-in replacement for TrackMate's regular LAP-Track algorithm.
Tracks out of the box for many types of datasets (bacteria, fluorescent nuclei, phase contrast cell culture etc), zero parameters to tune 🤖
www.molbiolcell.org/doi/full/10....

www.molbiolcell.org/doi/full/10....
The paper: arxiv.org/abs/2503.19545 1/🧵

The paper: arxiv.org/abs/2503.19545 1/🧵
My workflow will be to autotrace and then manually correct the autotraces, reducing the time for our tracing significantly!
My workflow will be to autotrace and then manually correct the autotraces, reducing the time for our tracing significantly!
📄 arxiv.org/pdf/2507.01009 (1/N)
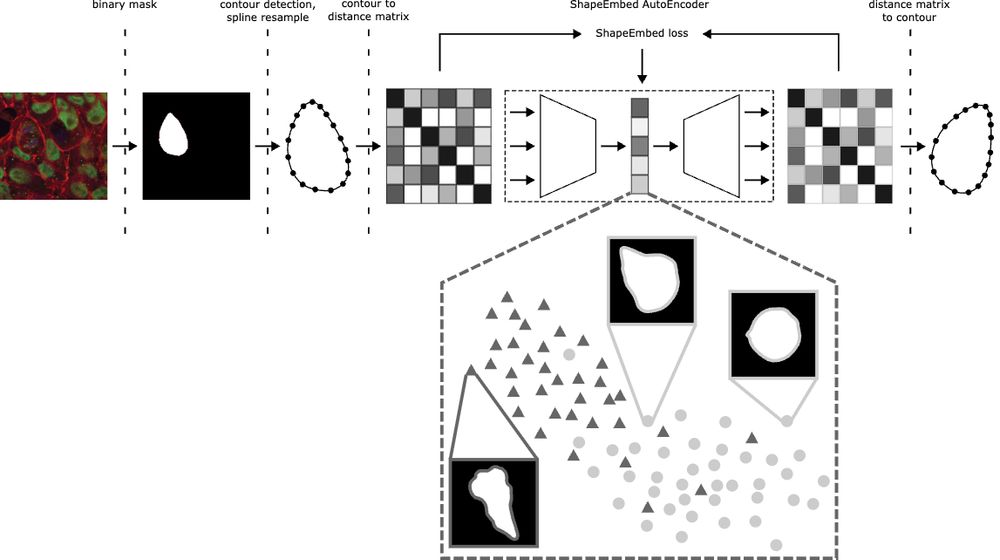
📄 arxiv.org/pdf/2507.01009 (1/N)
also shoutout to #microscopynodes by @aafkegros.bsky.social, here's to awesome new collabs born at blendercon!!!


www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
but @grosoane.bsky.social just added a very cool feature to toggle between color vision deficiencies. Watch your perceptually uniform color maps gain some kinks 😂
as a mild deutan myself, I appreciate it! 🙏
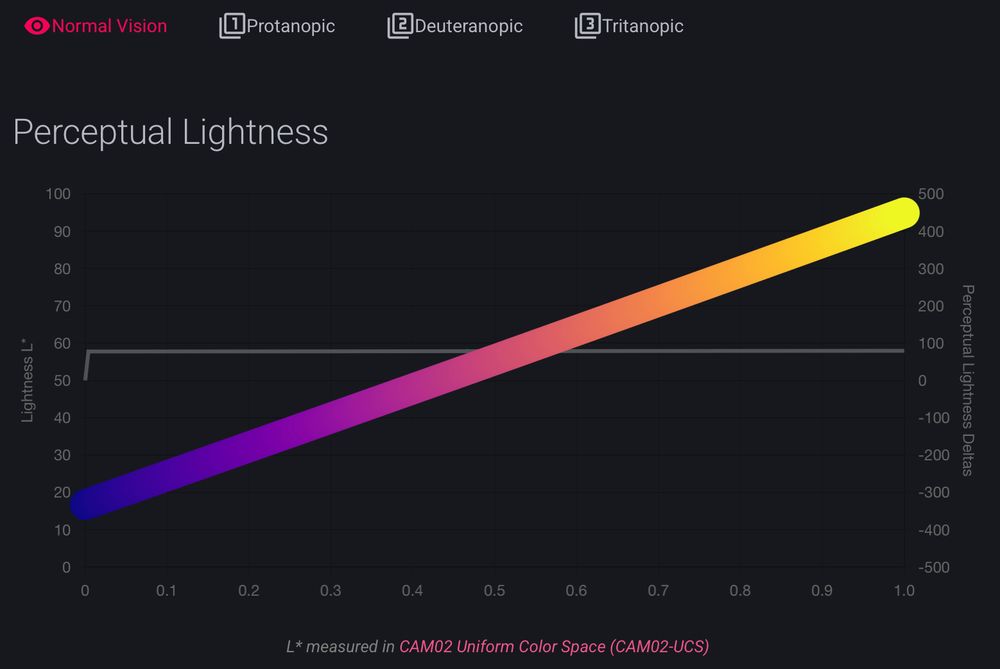
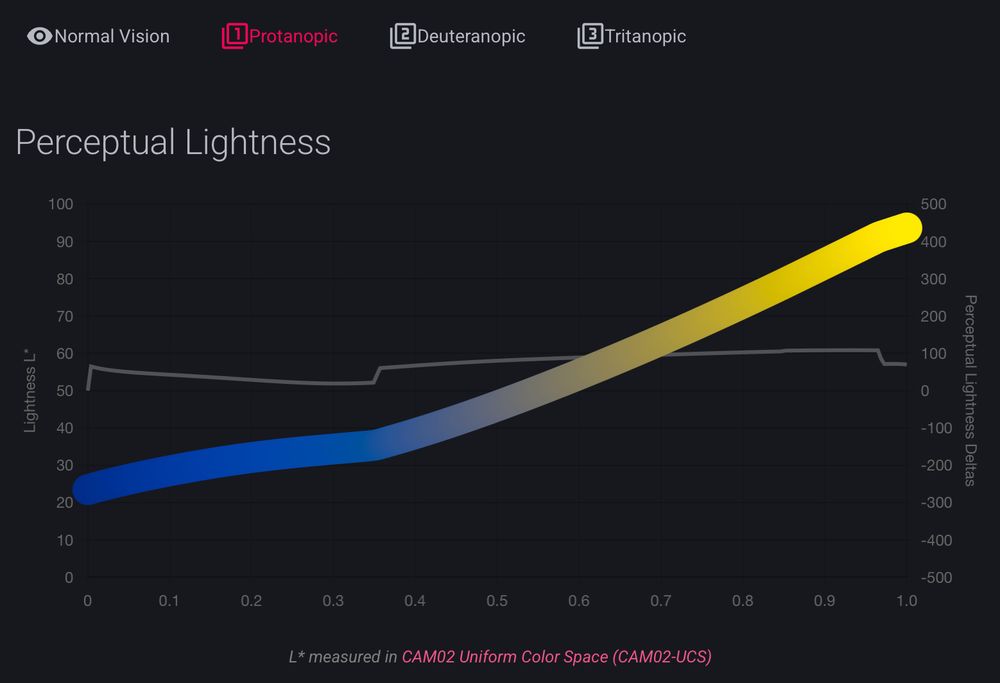
but @grosoane.bsky.social just added a very cool feature to toggle between color vision deficiencies. Watch your perceptually uniform color maps gain some kinks 😂
as a mild deutan myself, I appreciate it! 🙏
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
This packs lots of new fun features, including new color management 🌈, clearer transparency handling 🫥, custom default settings 🔧 and more!
Preprint at doi.org/10.1101/2025...
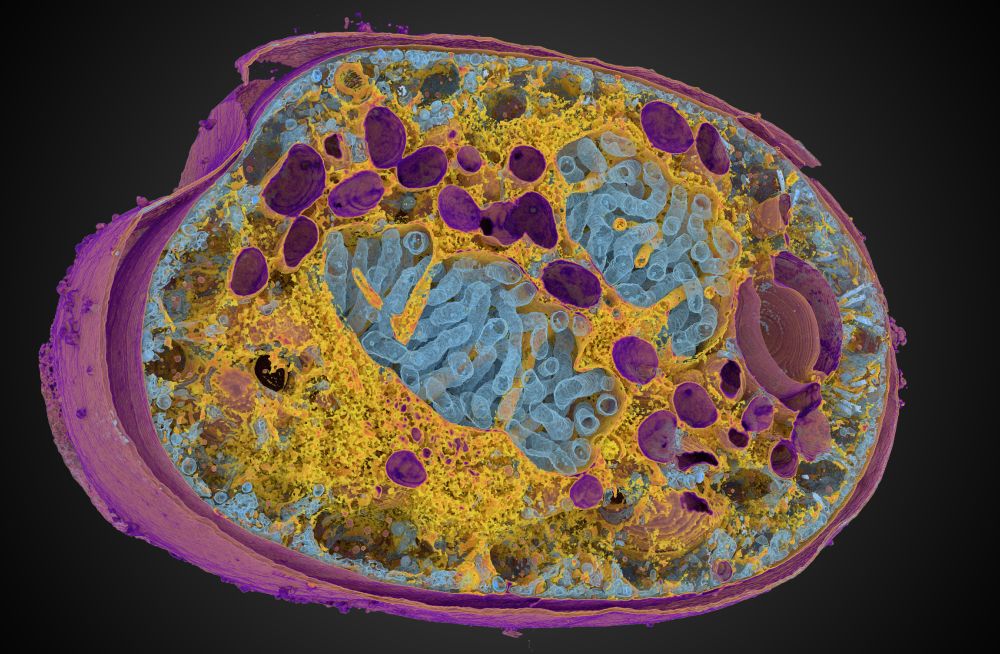
This packs lots of new fun features, including new color management 🌈, clearer transparency handling 🫥, custom default settings 🔧 and more!
Preprint at doi.org/10.1101/2025...
Release Highlights:
- Support for OME-zarr (v0.4)
- New workflow - Trainable Domain Adaptation
- New object features based on Spherical Harmonics Feature, contributed by @grosoane.bsky.social.
- Native support for Apple Silicon
Release Highlights:
- Support for OME-zarr (v0.4)
- New workflow - Trainable Domain Adaptation
- New object features based on Spherical Harmonics Feature, contributed by @grosoane.bsky.social.
- Native support for Apple Silicon
www.linkedin.com/posts/thermo...

www.youtube.com/watch?v=AYUy...

www.youtube.com/watch?v=AYUy...
Check it out here: academic.oup.com/nar/article/...
Check it out here: academic.oup.com/nar/article/...
Check any remote datasets (like @dudinlab.bsky.social maps of protists using ExM) in the highest quality.
For full details read forum.image.sc/t/mobie-plus...
Check any remote datasets (like @dudinlab.bsky.social maps of protists using ExM) in the highest quality.
For full details read forum.image.sc/t/mobie-plus...
Offers a bridge from Mastodon to Blender to create high quality 3D rendering of lineages and cell tracks over time:
(Tracking data of phallusia mammillata embryogenesis by Guignard et a. (2020). doi.org/10.1126/scie...)
Offers a bridge from Mastodon to Blender to create high quality 3D rendering of lineages and cell tracks over time:
(Tracking data of phallusia mammillata embryogenesis by Guignard et a. (2020). doi.org/10.1126/scie...)
This method quantifies the intensity distribution in microscopy objects, and is implemented parameter-free in @ilastik-team.bsky.social object classification!
We show applications in cells, C. elegans and Drosophila! 😁
This method quantifies the intensity distribution in microscopy objects, and is implemented parameter-free in @ilastik-team.bsky.social object classification!
We show applications in cells, C. elegans and Drosophila! 😁
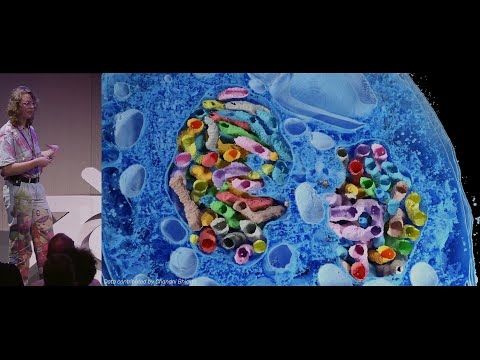
This is a Blender extension that seamlessly integrates and visualizes 3D microscopy data (TIF & @zarr.dev).
High-quality volume rendering for anyone, in both EM and fluorescence, regardless of computational expertise! 🔬
www.biorxiv.org/content/10.1...
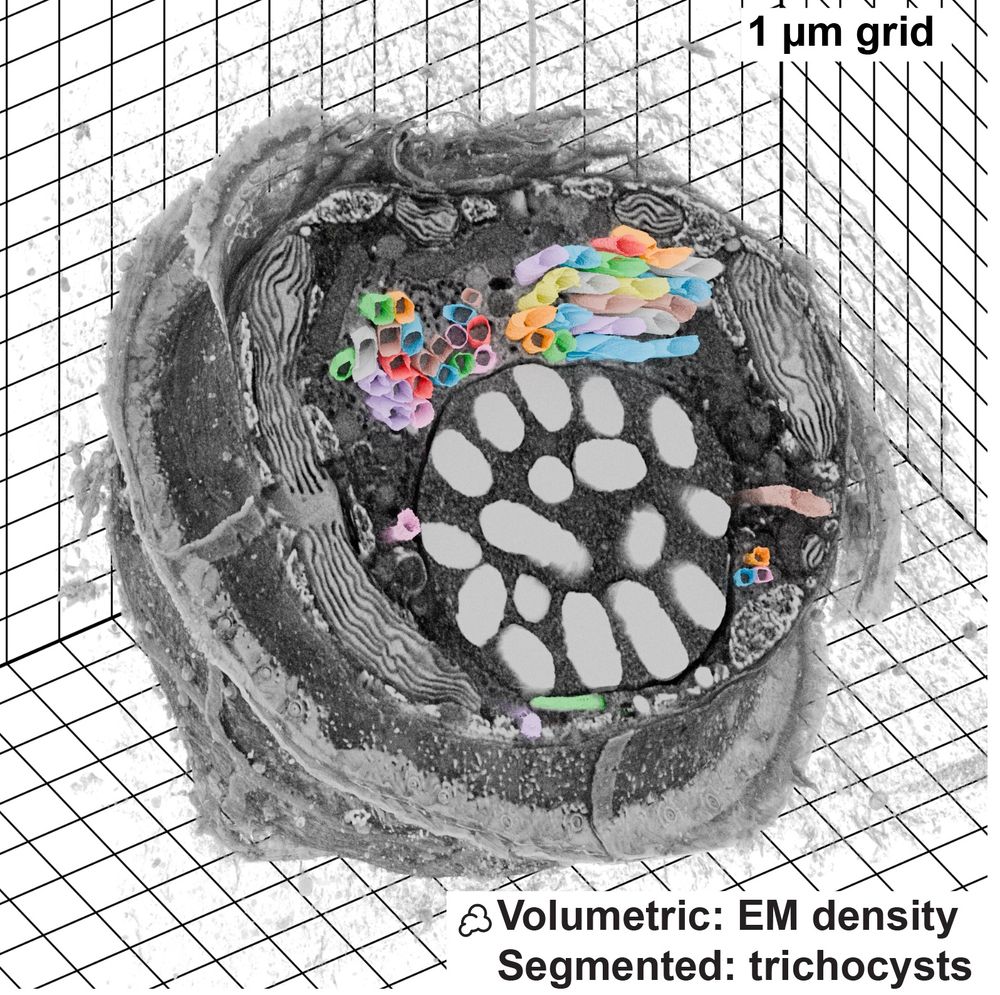
This is a Blender extension that seamlessly integrates and visualizes 3D microscopy data (TIF & @zarr.dev).
High-quality volume rendering for anyone, in both EM and fluorescence, regardless of computational expertise! 🔬
www.biorxiv.org/content/10.1...

