she/her | IPA: ˈafkə ɡrɔs
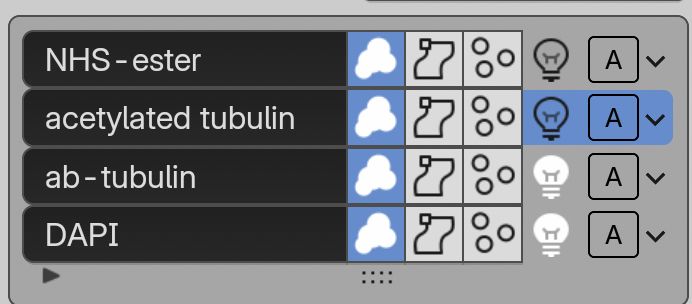
Following along is also easy with the example OME-Zarr @openmicroscopy.org datasets 😄
Find the tutorials here: www.youtube.com/playlist?lis...
Following along is also easy with the example OME-Zarr @openmicroscopy.org datasets 😄
Find the tutorials here: www.youtube.com/playlist?lis...
This packs lots of new fun features, including new color management 🌈, clearer transparency handling 🫥, custom default settings 🔧 and more!
Preprint at doi.org/10.1101/2025...
This packs lots of new fun features, including new color management 🌈, clearer transparency handling 🫥, custom default settings 🔧 and more!
Preprint at doi.org/10.1101/2025...
Here we can see how the channel distributions all align in the projection after we optically activate Rac1 in one part of the cell. 💡
With @jpassmore.bsky.social , Lukas Kapitein
Here we can see how the channel distributions all align in the projection after we optically activate Rac1 in one part of the cell. 💡
With @jpassmore.bsky.social , Lukas Kapitein
We show this for classification of meiotic zones in C. elegans :) 🪱
For the nerds there's also a Python API 😉
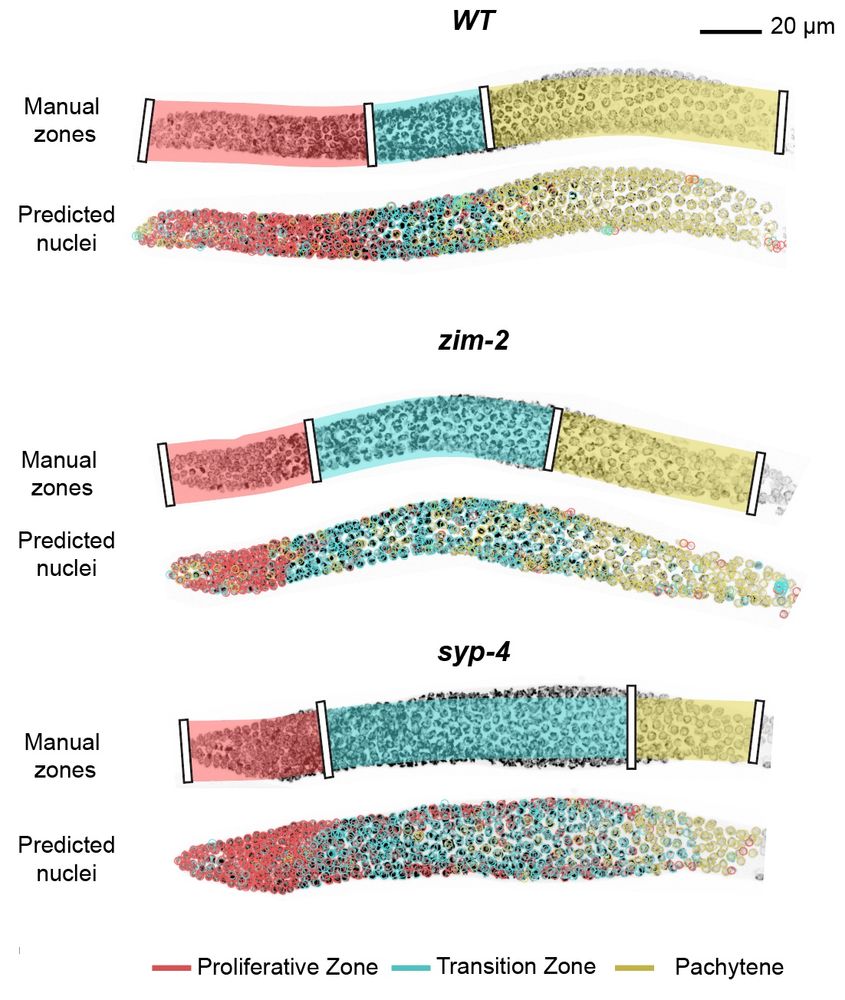
We show this for classification of meiotic zones in C. elegans :) 🪱
For the nerds there's also a Python API 😉
In collaboration with
@ottilie.bsky.social, @timothyfuqua.bsky.social, @justinmcrocker.bsky.social
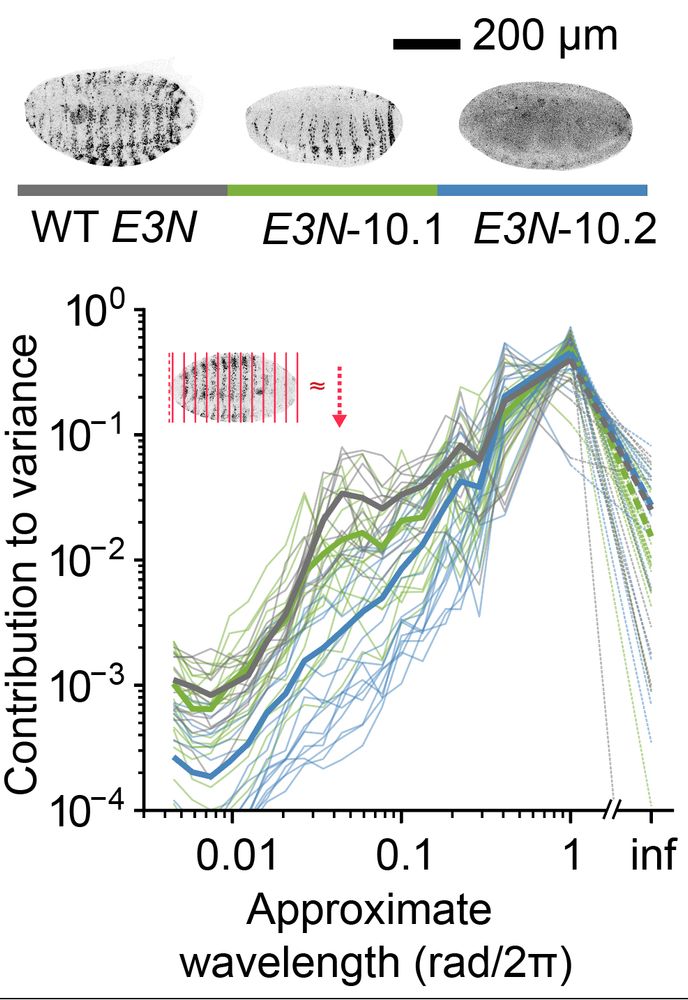
In collaboration with
@ottilie.bsky.social, @timothyfuqua.bsky.social, @justinmcrocker.bsky.social
This gives us a metric that shows how detailed the features in the spherical map are, giving a rotation-invariant quantification of distribution.
This gives us a metric that shows how detailed the features in the spherical map are, giving a rotation-invariant quantification of distribution.
This video is made with @blender3d.bsky.social geometry nodes and Microscopy Nodes, allowing the microscopy and method illustration in the same video :D
This video is made with @blender3d.bsky.social geometry nodes and Microscopy Nodes, allowing the microscopy and method illustration in the same video :D
This method quantifies the intensity distribution in microscopy objects, and is implemented parameter-free in @ilastik-team.bsky.social object classification!
We show applications in cells, C. elegans and Drosophila! 😁
This method quantifies the intensity distribution in microscopy objects, and is implemented parameter-free in @ilastik-team.bsky.social object classification!
We show applications in cells, C. elegans and Drosophila! 😁
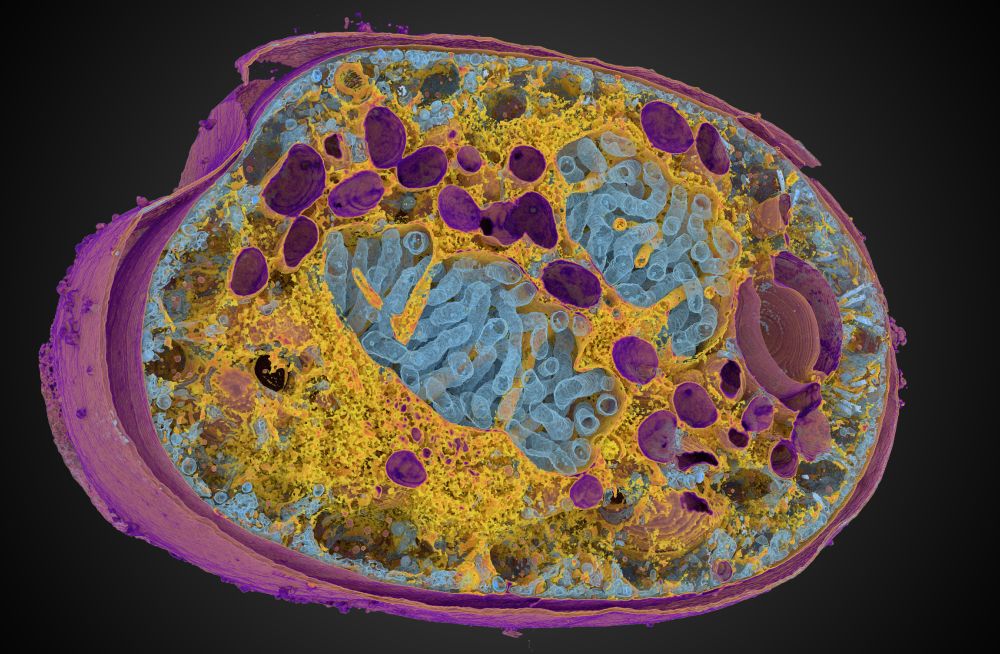
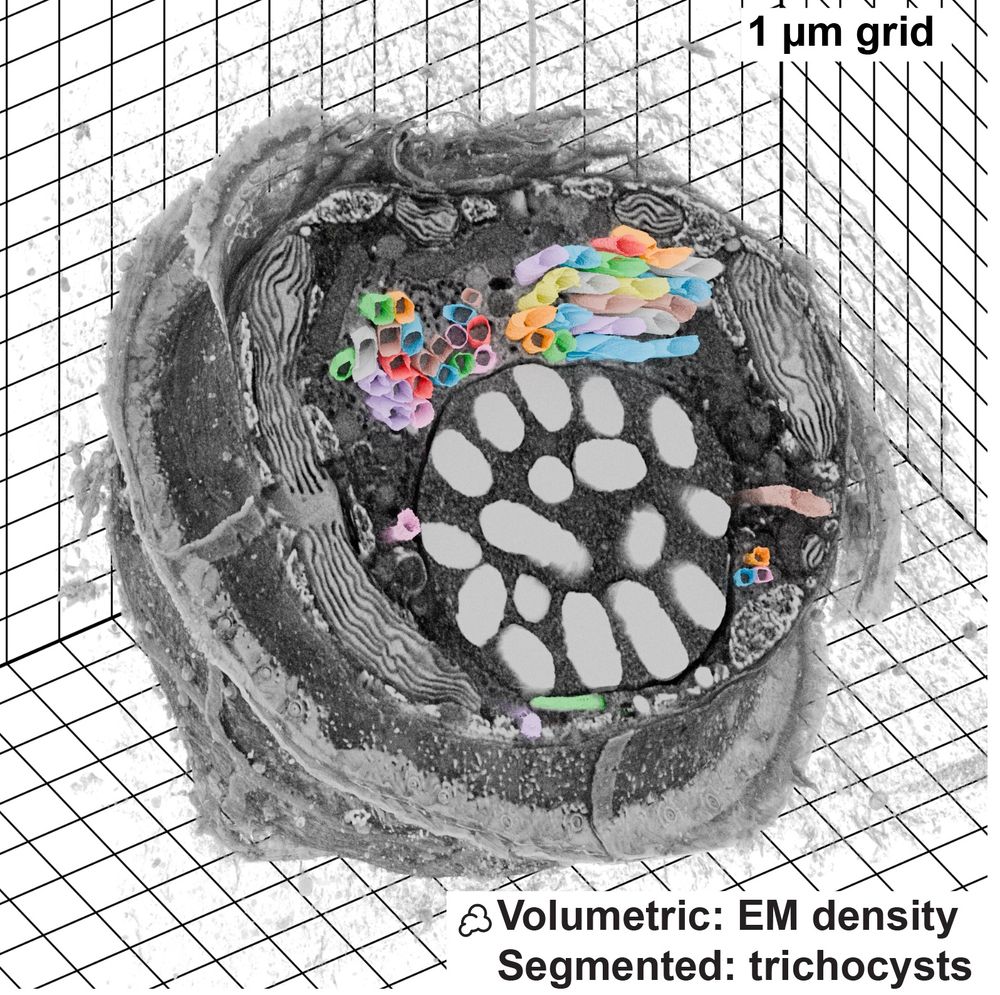
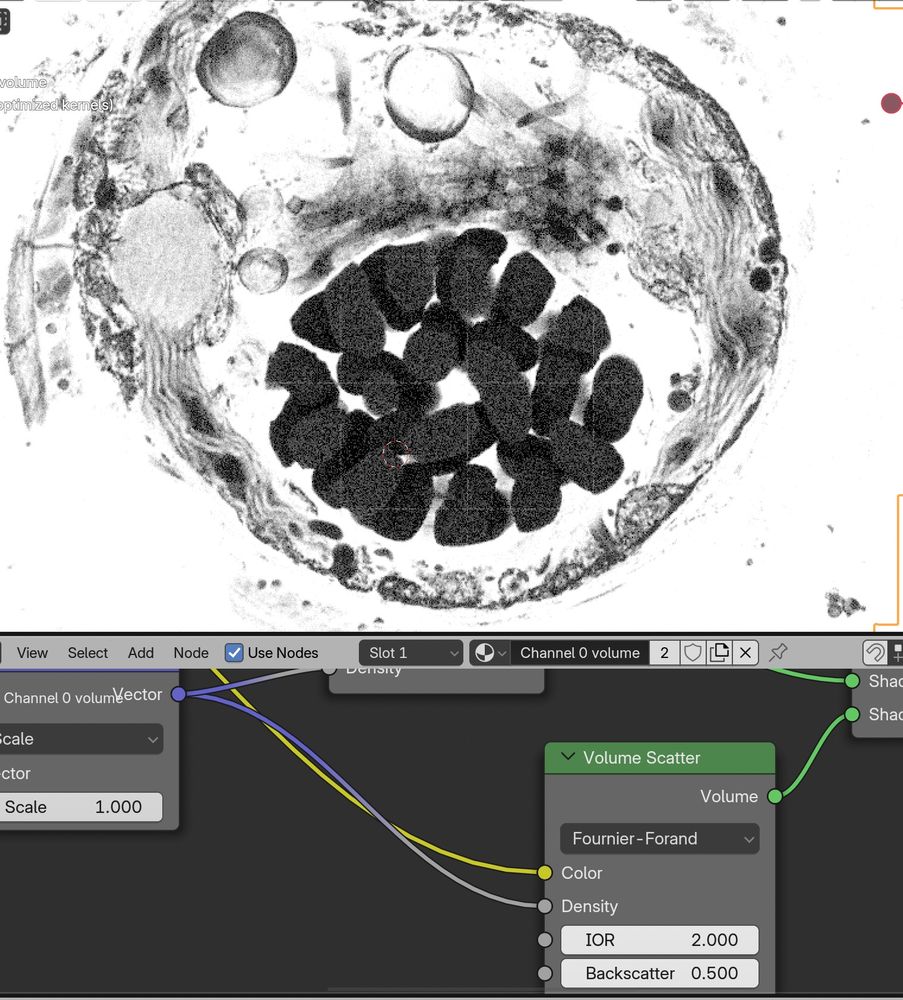
The data here is a dinoflagellate from Mocaer et al. 2023, hosted publicly at EMPIAR-11399 (also shown in the first post here).
The data here is a dinoflagellate from Mocaer et al. 2023, hosted publicly at EMPIAR-11399 (also shown in the first post here).
This is a mitotic cell from Walther et al. 2018, that's hosted publicly on the Image Data Resource 😱
This is a mitotic cell from Walther et al. 2018, that's hosted publicly on the Image Data Resource 😱
With the sparse volume render in Blender you can see the network of tunnels through the nucleus that organize the spindle in its closed mitosis! 🤩
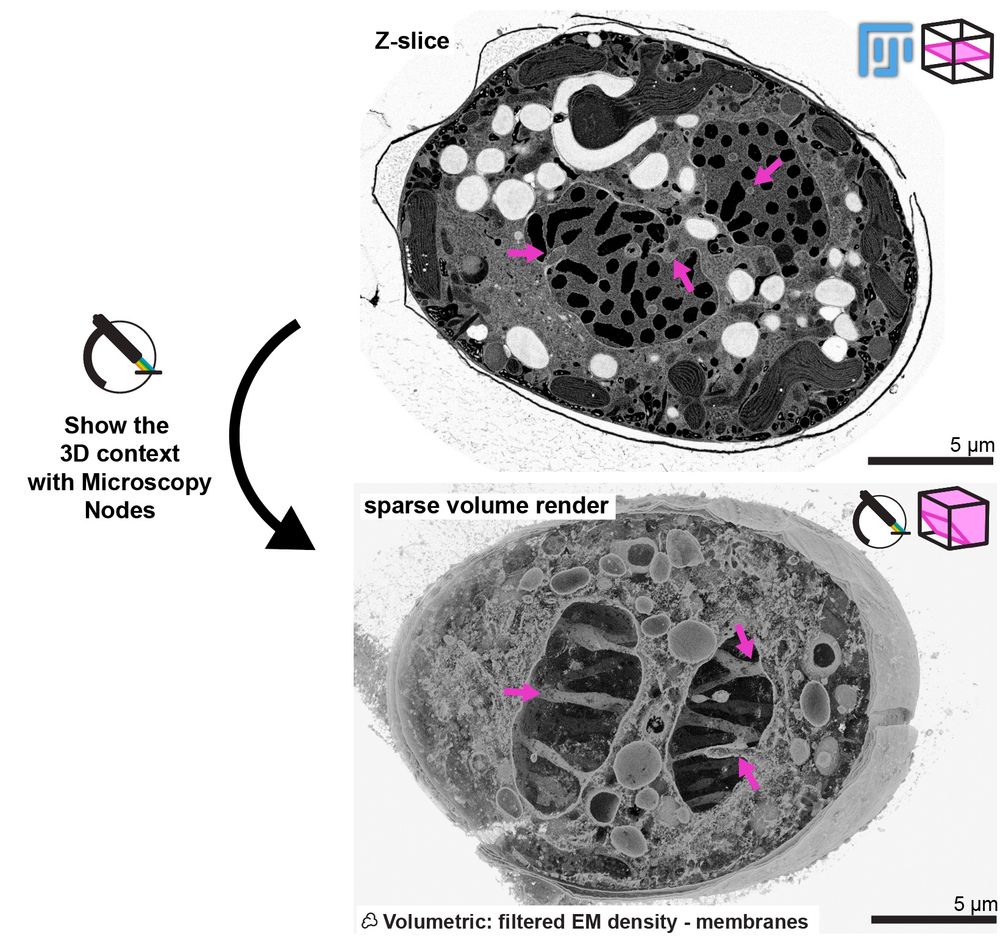
With the sparse volume render in Blender you can see the network of tunnels through the nucleus that organize the spindle in its closed mitosis! 🤩
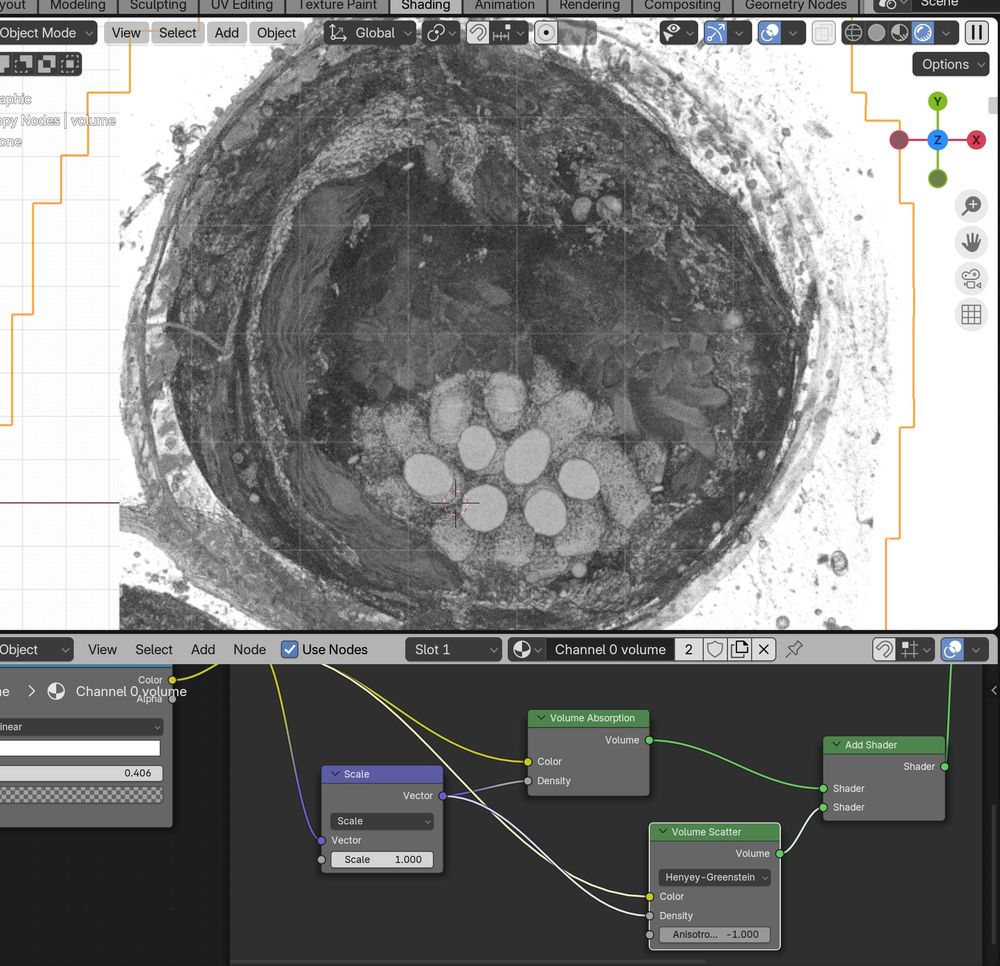
This is a Blender extension that seamlessly integrates and visualizes 3D microscopy data (TIF & @zarr.dev).
High-quality volume rendering for anyone, in both EM and fluorescence, regardless of computational expertise! 🔬
www.biorxiv.org/content/10.1...
This is a Blender extension that seamlessly integrates and visualizes 3D microscopy data (TIF & @zarr.dev).
High-quality volume rendering for anyone, in both EM and fluorescence, regardless of computational expertise! 🔬
www.biorxiv.org/content/10.1...

Great info and work on the upcoming features of Zarr here :D Transforms, collections, and more! 🙏
Great info and work on the upcoming features of Zarr here :D Transforms, collections, and more! 🙏
(data from Granita Lokaj, @ s3.embl.de/microscopyno...)

(data from Granita Lokaj, @ s3.embl.de/microscopyno...)