
Professor of Molecular Systems Biology, University of Cambridge

Congrats Hannah Ochner and authors on this important paper! Strong collaboration with @kiranrpatil.bsky.social
www.biorxiv.org/cgi/content/...
@mrclmb.bsky.social @wellcometrust.bsky.social
#science #microbiome #health
www.nature.com/articles/s41...
Reposted by Kiran Raosaheb Patil

#science #microbiome #health
www.nature.com/articles/s41...
Reposted by Kiran Raosaheb Patil





www.nature.com/articles/s41...
Reposted by Kiran Raosaheb Patil
🚨Out now!
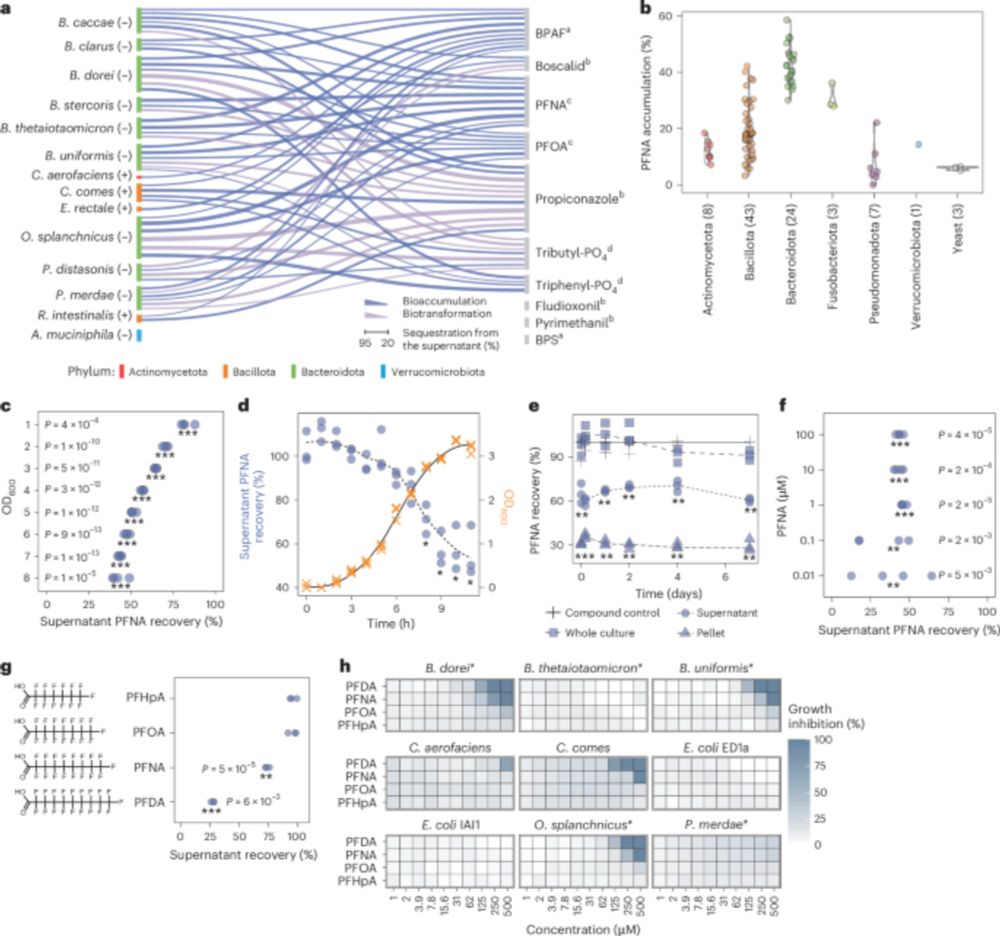
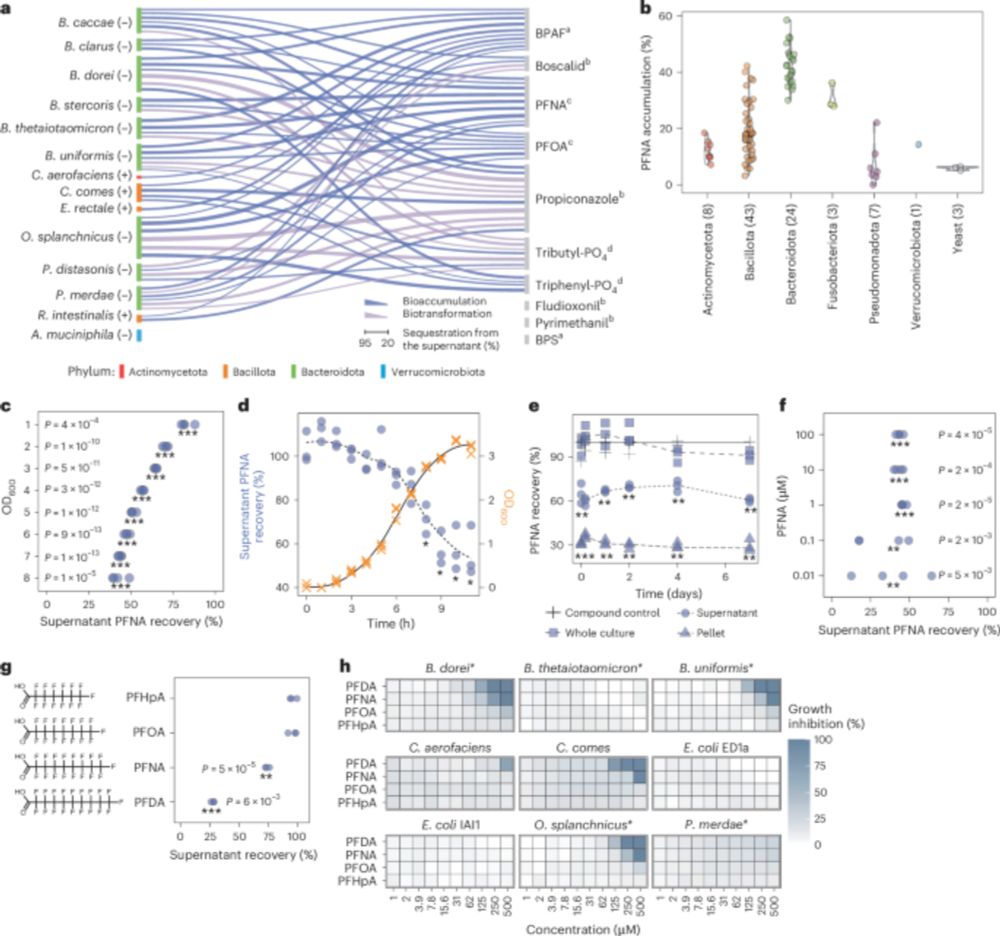
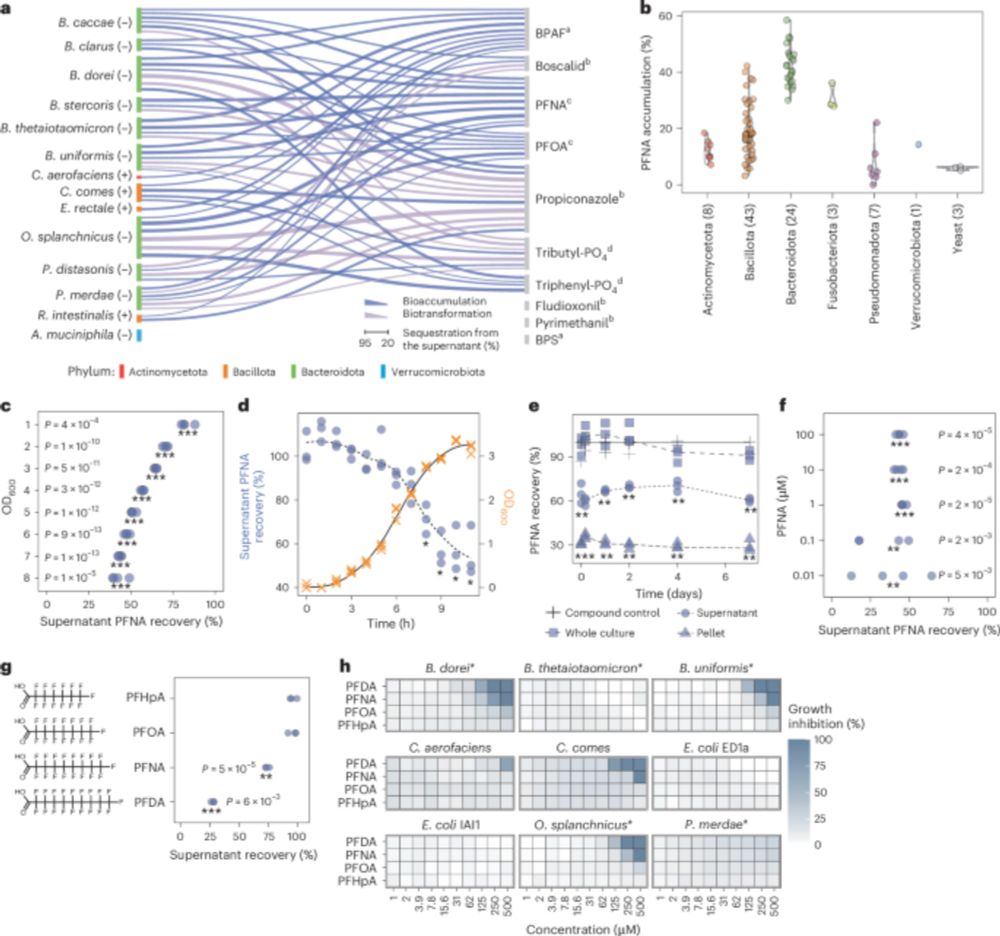
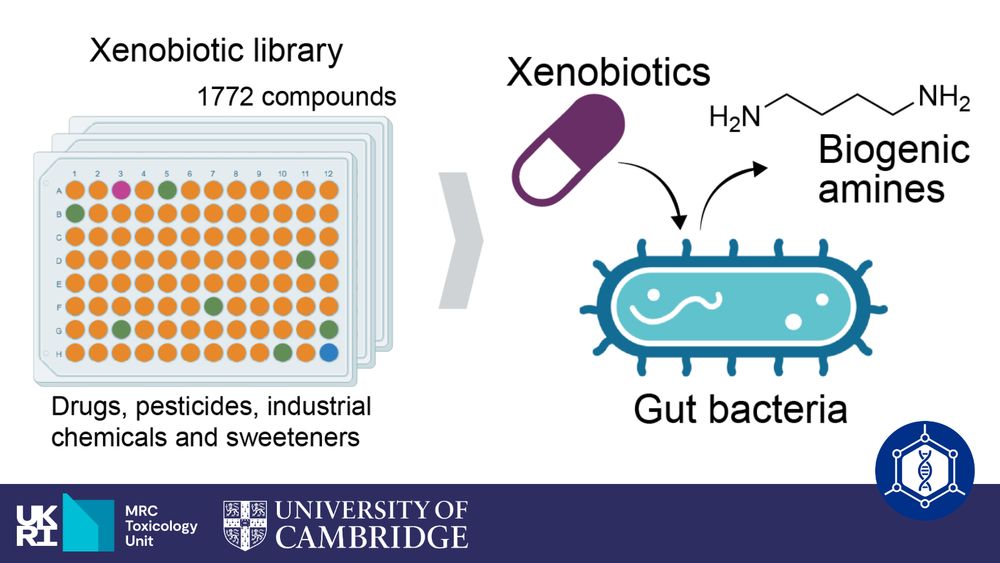
Human gut bacteria bioaccumulate forever chemicals, in intracellular aggregates and colonization of gnotobiotic mice with these bioaccumulating bacteria increases faecal PFAS excretion. @kiranrpatil.bsky.social
#MicroSky #MicrobiomeSky 🦠
www.nature.com/articles/s41...

Reposted by Kiran Raosaheb Patil
Reposted by Kiran Raosaheb Patil
Reposted by Michael Wagner, Kiran Raosaheb Patil
They can shield us from harmful chemicals—and the latest study from Kiran Patil's lab (@kiranrpatil.bsky.social)
uncovers how. We’re proud to have contributed to this exciting work!
#Microbiome #Detox
www.nature.com/articles/s41...
Reposted by Kiran Raosaheb Patil
Read more here: buff.ly/4FsVFsS
@kiranrpatil.bsky.social @indraroux.bsky.social
Find out more here: buff.ly/5akzsgs
@kiranrpatil.bsky.social @skamrad.bsky.social
Reposted by Kiran Raosaheb Patil

Find out more here: buff.ly/5akzsgs
@kiranrpatil.bsky.social @skamrad.bsky.social
Reposted by Kiran Raosaheb Patil

We systematically map proteomic and metabolomic interactions between >100 pairs of gut bacteria, detecting metabolic interactions and functionally-related clusters of proteins.
www.biorxiv.org/content/10.1...
@kiranrpatil.bsky.social
Reposted by Kiran Raosaheb Patil

We show how glycolytic activity instructs germ layer proportions through regulation of Nodal and Wnt signaling - happy to finally share this 😊
doi.org/10.1016/j.st...
B2B with @jesseveenvliet.bsky.social lab: doi.org/10.1016/j.st... 🤩
@kiranrpatil.bsky.social:
Obligate cross-feeding of metabolites is common in soil microbial communities
By Ghada Yousif @metagenomez.bsky.social with @swagatika.bsky.social @isamirgiri.bsky.social Sharvari Harshe et al.
www.biorxiv.org/content/10.1...
🧵👇

Reposted by Kiran Raosaheb Patil

@kiranrpatil.bsky.social:
Obligate cross-feeding of metabolites is common in soil microbial communities
By Ghada Yousif @metagenomez.bsky.social with @swagatika.bsky.social @isamirgiri.bsky.social Sharvari Harshe et al.
www.biorxiv.org/content/10.1...
🧵👇



