
n-way operations on tensors a la parafac etc
They are virtually the same in spectral hemistry, neuro (EEG, fMRI), BI, psychology etc all rare differently named etc
There is plenty more as we lack ability to properly search by "meaning" not name or semantics
Custom embeddings might help
n-way operations on tensors a la parafac etc
They are virtually the same in spectral hemistry, neuro (EEG, fMRI), BI, psychology etc all rare differently named etc
There is plenty more as we lack ability to properly search by "meaning" not name or semantics
Custom embeddings might help
FCNCP: A Coupled Nonnegative CANDECOMP/PARAFAC Decomposition Based on Federated Learning
https://arxiv.org/abs/2404.11890
FCNCP: A Coupled Nonnegative CANDECOMP/PARAFAC Decomposition Based on Federated Learning
https://arxiv.org/abs/2404.11890


2/3
2/3
Kristina P. Sinaga
http://arxiv.org/abs/2509.16101

Kristina P. Sinaga
http://arxiv.org/abs/2509.16101

Abstract: Tensor CANDECOMP/PARAFAC decomposition (CPD) is a fundamental model for tensor reconstruction. Although the Bayesian framework allows for principled uncertainty quantification and automatic [1/4 of https://arxiv.org/abs/2505.16305v1]
Abstract: Tensor CANDECOMP/PARAFAC decomposition (CPD) is a fundamental model for tensor reconstruction. Although the Bayesian framework allows for principled uncertainty quantification and automatic [1/4 of https://arxiv.org/abs/2505.16305v1]
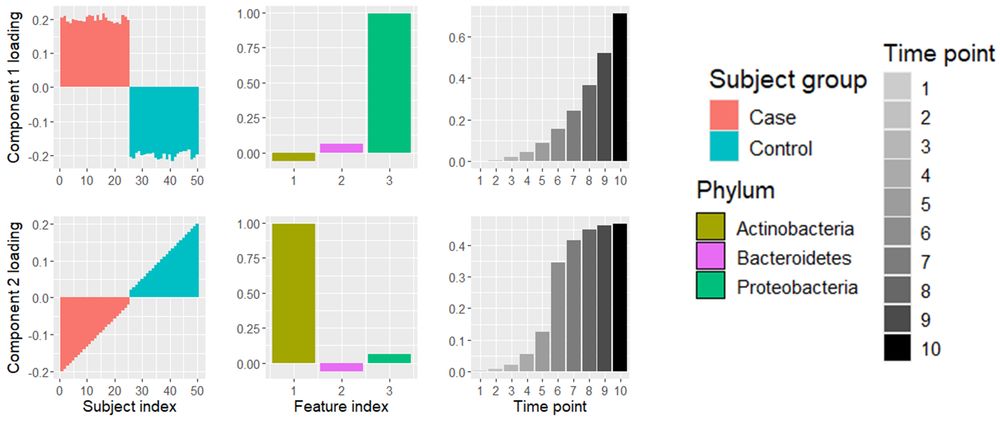
Latent Functional PARAFAC for modeling multidimensional longitudinal data
https://arxiv.org/abs/2410.18696
Latent Functional PARAFAC for modeling multidimensional longitudinal data
https://arxiv.org/abs/2410.18696
#AcademicTwitter #rStats

#AcademicTwitter #rStats
Biplots are a very useful (and underused) tool to visualise interactions.
1/3
Biplots are a very useful (and underused) tool to visualise interactions.
1/3

