
www.biorxiv.org/content/10.1...
Read below for a few highlights...
We reveal a remarkable cell type diversity, nerve net condensation, a multilayered circuit & gene expression profiles.
www.biorxiv.org/content/10.1... #ctenophores
We reveal a remarkable cell type diversity, nerve net condensation, a multilayered circuit & gene expression profiles.
www.biorxiv.org/content/10.1... #ctenophores
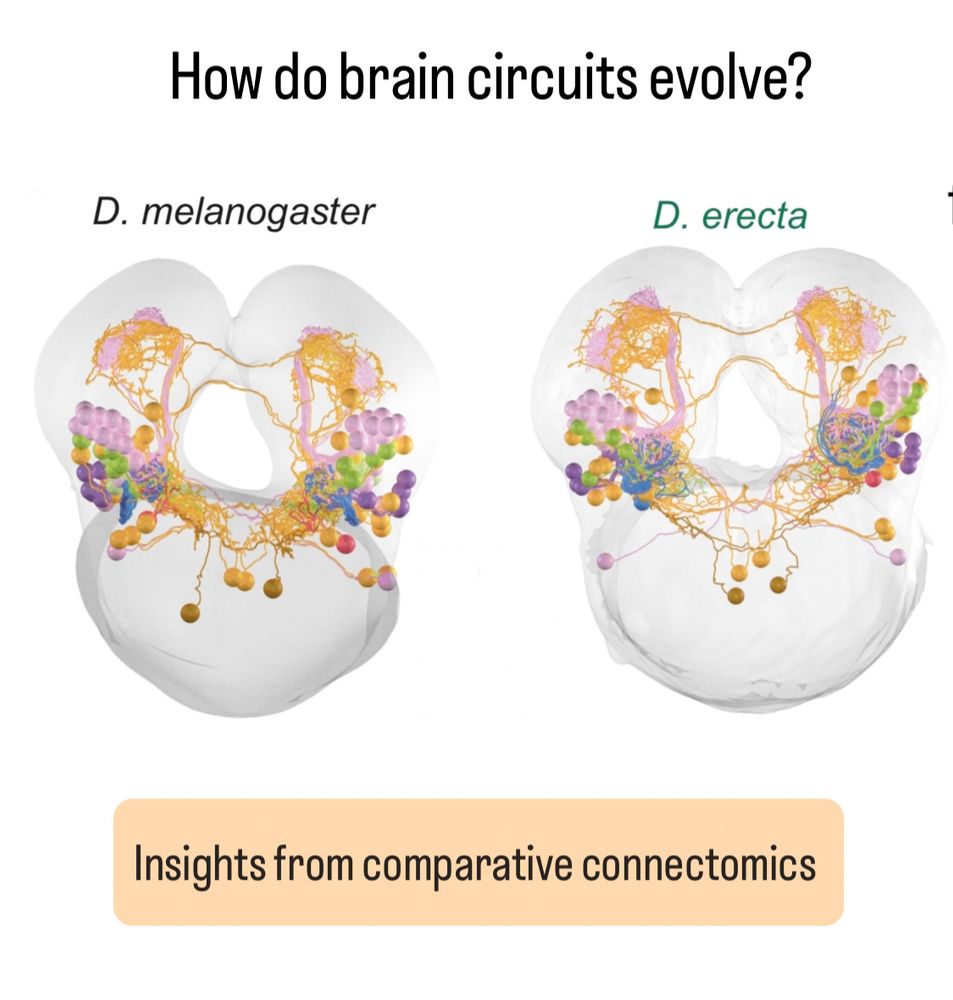
We explored its remarkable cellular diversity and integration with the syncytial nerve net, uncovering connectivity and insights into the evolution of sensory complexes.
www.biorxiv.org/content/10.1...

We explored its remarkable cellular diversity and integration with the syncytial nerve net, uncovering connectivity and insights into the evolution of sensory complexes.
www.biorxiv.org/content/10.1...
www.cell.com/cell/fulltex...

www.cell.com/cell/fulltex...
bit.ly/44aVm9E
bit.ly/44aVm9E
Roscoff feels like a personal refugium - a timeless place where one’s soul can hide & think about things that really matter.



Roscoff feels like a personal refugium - a timeless place where one’s soul can hide & think about things that really matter.
Lineage-resolved analysis of embryonic gene expression evolution in C. elegans and C. briggsae | Science www.science.org/doi/10.1126/...
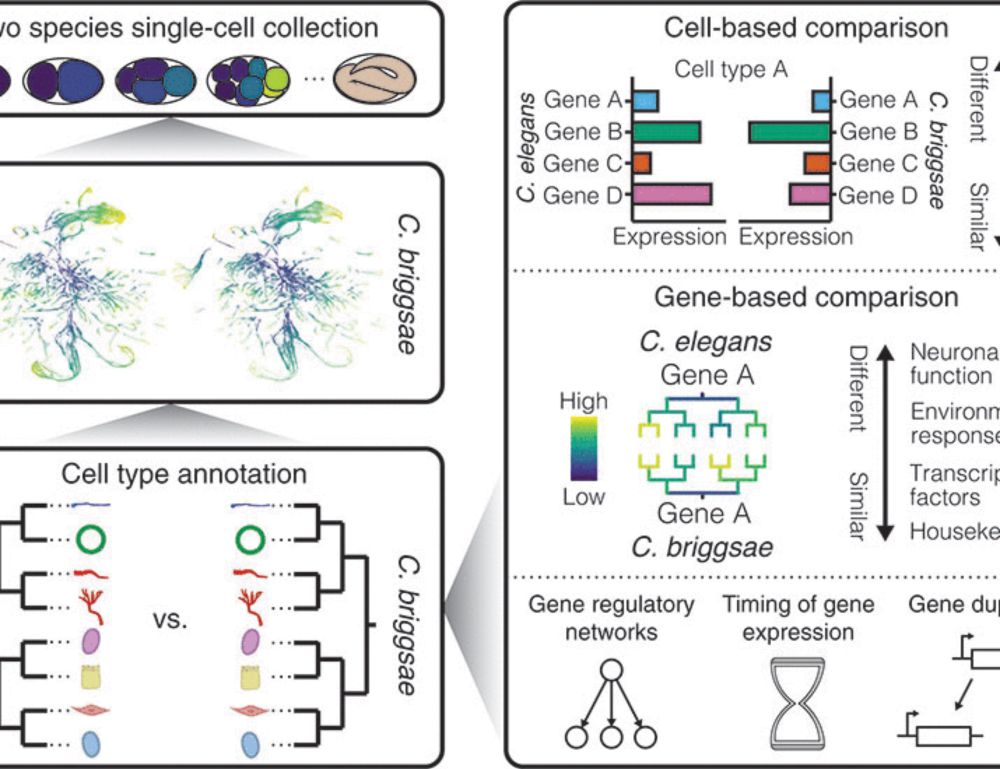
Lineage-resolved analysis of embryonic gene expression evolution in C. elegans and C. briggsae | Science www.science.org/doi/10.1126/...
@ruthstyfhals.bsky.social

@ruthstyfhals.bsky.social


... The Woodstock of Biology2 & Night Science was terminated by police arrival.
>The End<
Nobody apparently dared to take a picture but it's true.


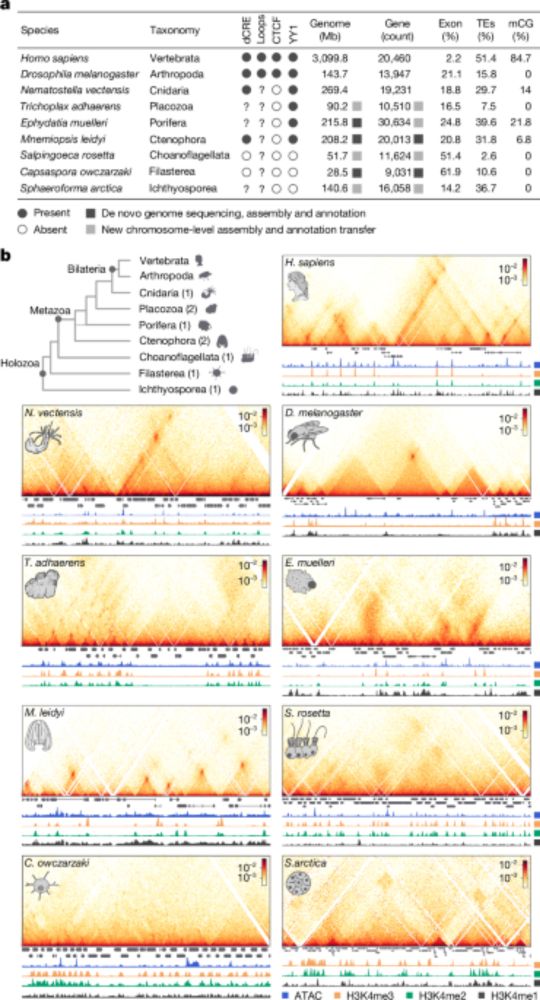
Lots of interesting new TE (& genes) biology
Data fully browsable💻 👉 embryo.helmholtz-munich.de/shiny_embryo/
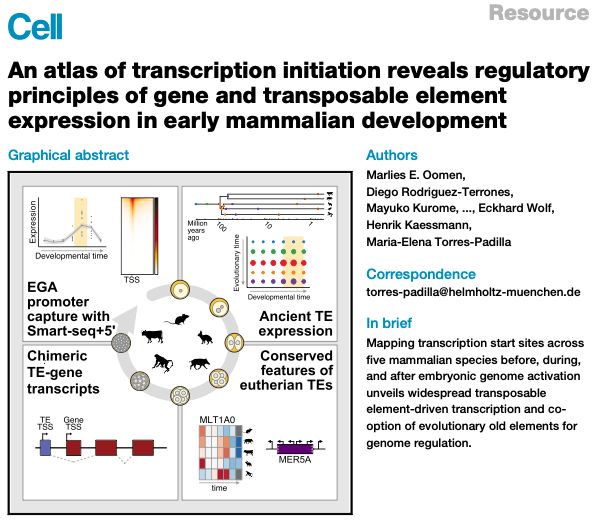
Lots of interesting new TE (& genes) biology
Data fully browsable💻 👉 embryo.helmholtz-munich.de/shiny_embryo/
Thanks, @fwf-at.bsky.social for funding our work!
bmcbiol.biomedcentral.com/articles/10....
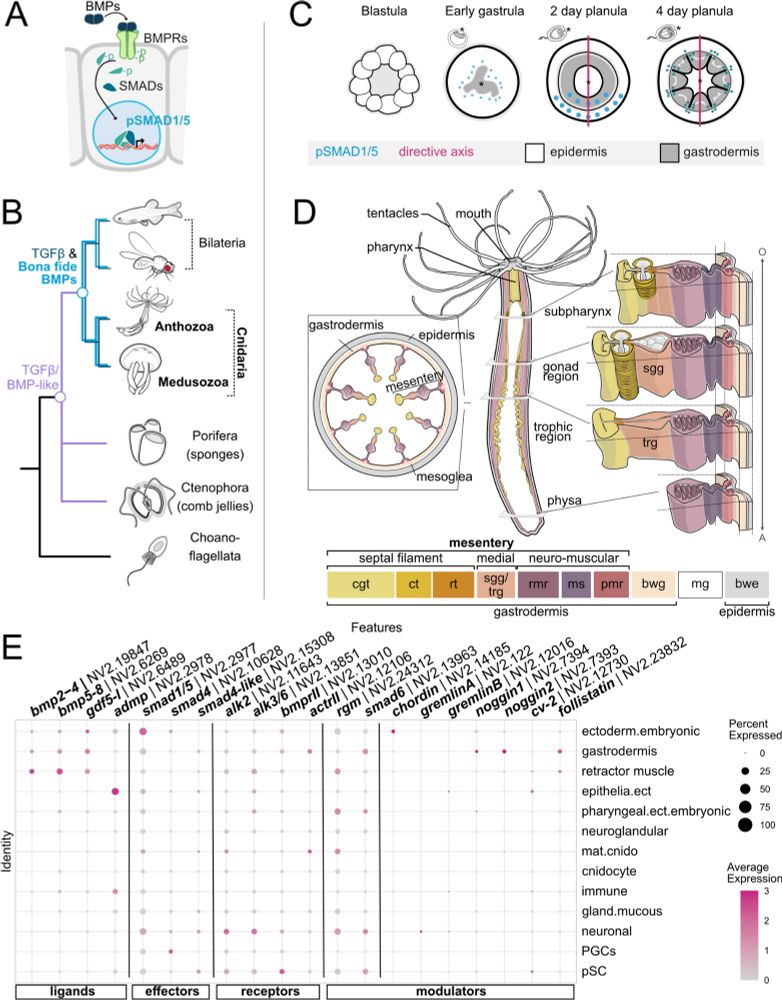
Thanks, @fwf-at.bsky.social for funding our work!
bmcbiol.biomedcentral.com/articles/10....
people manage to get babies in this timeframe.

people manage to get babies in this timeframe.
www.nature.com/articles/s41...
Unfortunately, it makes several misleading claims that we disagree with. We will address these technical & other issues with this work over the next few months.

www.nature.com/articles/s41...
Unfortunately, it makes several misleading claims that we disagree with. We will address these technical & other issues with this work over the next few months.
https://buff.ly/4jnWk82

https://buff.ly/4jnWk82





