Working on data structures and algorithms for pangenome graphs

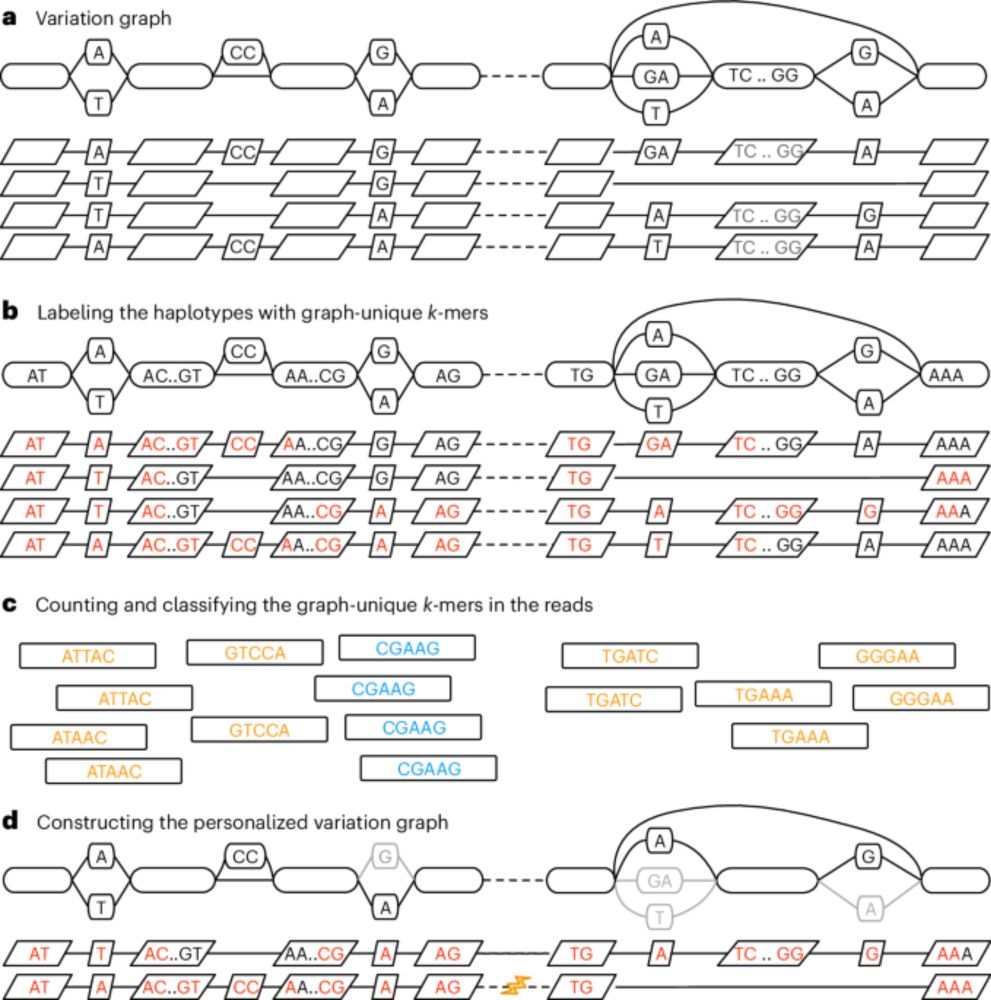
Mapping long reads to pangenome graphs is ~10x faster than with GraphAligner, with veeery slightly better mapping accuracy, short variant calling, and SV genotyping than GraphAligner or Minimap2
Mapping long reads to pangenome graphs is ~10x faster than with GraphAligner, with veeery slightly better mapping accuracy, short variant calling, and SV genotyping than GraphAligner or Minimap2

Keep an eye on this space for updates, event information, and ways to get involved. We can't wait to see everyone #standupforscience2025 on March 7th, both in DC and locations nationwide!
#scienceforall #sciencenotsilence

Keep an eye on this space for updates, event information, and ways to get involved. We can't wait to see everyone #standupforscience2025 on March 7th, both in DC and locations nationwide!
#scienceforall #sciencenotsilence
🗞 news.ucsc.edu/2025/01/pate...
👏🏼👏🏿👏🏾 @khmiga.bsky.social @benedictpaten.bsky.social

🗞 news.ucsc.edu/2025/01/pate...
👏🏼👏🏿👏🏾 @khmiga.bsky.social @benedictpaten.bsky.social


