Wei-Ting Lu
@weitinglu.bsky.social
Tea addict and scientist, in that order. Incoming Group Leader, Dept of Oncology, University of Oxford. All views my own.
Reposted by Wei-Ting Lu
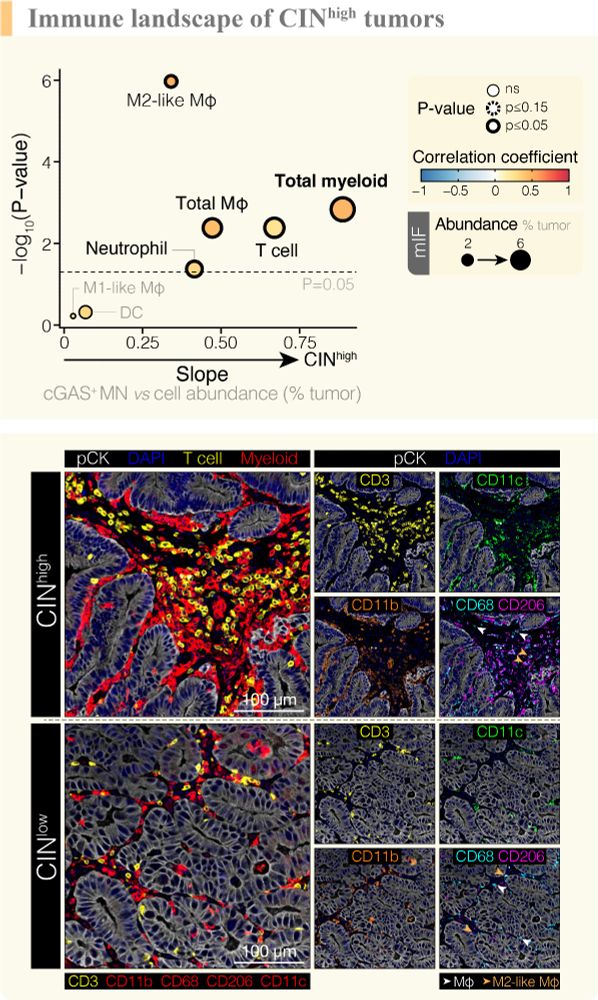
Using snRNA-sequencing (Izar lab @columbiauniversity.bsky.social) with matched multiplex immunophenotypic profiling (@jleslie1.bsky.social) we found tumor cell-intrinsic innate immune activation and intratumoral myeloid infiltration as phenotypic consequences of ongoing micronucleation in EAC. 7/11
May 12, 2025 at 12:08 PM
Using snRNA-sequencing (Izar lab @columbiauniversity.bsky.social) with matched multiplex immunophenotypic profiling (@jleslie1.bsky.social) we found tumor cell-intrinsic innate immune activation and intratumoral myeloid infiltration as phenotypic consequences of ongoing micronucleation in EAC. 7/11
Reposted by Wei-Ting Lu
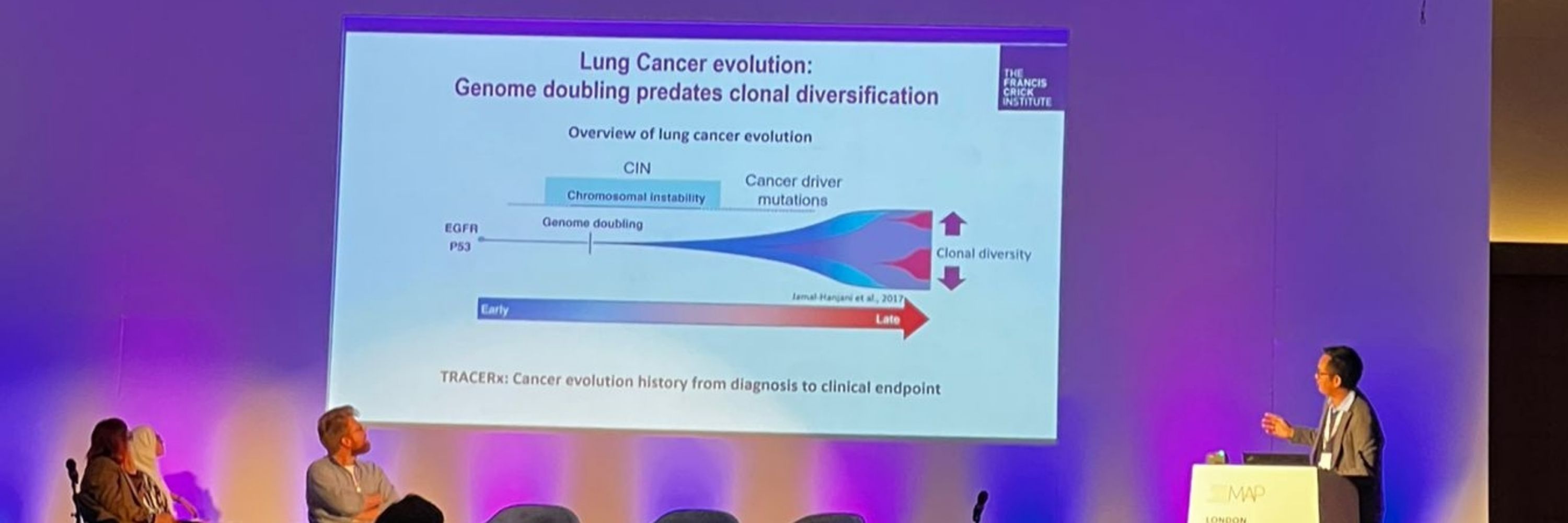
Wei-ting Lu presents data from the TRACERx lung cancer study on FAT1 alterations and chromosomal instability.
#AACR2025 | @weitinglu.bsky.social | @crick.ac.uk
#AACR2025 | @weitinglu.bsky.social | @crick.ac.uk
April 27, 2025 at 3:33 PM
Wei-ting Lu presents data from the TRACERx lung cancer study on FAT1 alterations and chromosomal instability.
#AACR2025 | @weitinglu.bsky.social | @crick.ac.uk
#AACR2025 | @weitinglu.bsky.social | @crick.ac.uk
I got an idea to make you feel *worse* 🙃🙃-- read Michael Lewis's the fifth risk.
January 30, 2025 at 6:14 PM
I got an idea to make you feel *worse* 🙃🙃-- read Michael Lewis's the fifth risk.
We are fortunate to get a trial of the NL5+ confocal system from confocal.nl which is a total gamechanger. @crick.ac.uk @calm-stp.bsky.social just got a NL5+ system after the trial -- can't wait to have more fun with it♥️!! 4/4
Homepage - Confocal NL - Designed to respect your cells
Discover the confocal systems that enable you to perform deep and fast live cell imaging, beyond the diffraction limit using only nanowatts of laser power.
confocal.nl
January 17, 2025 at 3:17 PM
We are fortunate to get a trial of the NL5+ confocal system from confocal.nl which is a total gamechanger. @crick.ac.uk @calm-stp.bsky.social just got a NL5+ system after the trial -- can't wait to have more fun with it♥️!! 4/4
The biggest nemesis of live microscopy is light toxicity. The success rate for mitotic imaging is painfully low because one has to balance resolution vs cell death – either you have a blurry movie or a high-resolution movie showing a dead cell. 3/4
January 17, 2025 at 3:17 PM
The biggest nemesis of live microscopy is light toxicity. The success rate for mitotic imaging is painfully low because one has to balance resolution vs cell death – either you have a blurry movie or a high-resolution movie showing a dead cell. 3/4
The cover is designed by Jeroen at Phsopho Animation,
who converted a mitotis time-lapse movie into some awesome cover art. One can see that chromosomes (white) align and then pulled apart by tubulin (red) as the cell divides, showing lagging chromosomes and chromosomal bridges. 2/4
who converted a mitotis time-lapse movie into some awesome cover art. One can see that chromosomes (white) align and then pulled apart by tubulin (red) as the cell divides, showing lagging chromosomes and chromosomal bridges. 2/4
January 17, 2025 at 3:17 PM
The cover is designed by Jeroen at Phsopho Animation,
who converted a mitotis time-lapse movie into some awesome cover art. One can see that chromosomes (white) align and then pulled apart by tubulin (red) as the cell divides, showing lagging chromosomes and chromosomal bridges. 2/4
who converted a mitotis time-lapse movie into some awesome cover art. One can see that chromosomes (white) align and then pulled apart by tubulin (red) as the cell divides, showing lagging chromosomes and chromosomal bridges. 2/4
We hope this is useful for the field. Our paper can be found here:
www.nature.com/articles/s41...
Or the readcube link below:
rdcu.be/d5gAe
Finally we wish everyone a happy new year!!! 14/end
www.nature.com/articles/s41...
Or the readcube link below:
rdcu.be/d5gAe
Finally we wish everyone a happy new year!!! 14/end
TRACERx analysis identifies a role for FAT1 in regulating chromosomal instability and whole-genome doubling via Hippo signalling - Nature Cell Biology
Lu et al. perform systematic functional analyses using data from the TRACERx cohort of patients with non-small-cell lung cancer and delineate how FAT1 regulates homologous recombination repair, chromo...
www.nature.com
January 3, 2025 at 11:24 AM
We hope this is useful for the field. Our paper can be found here:
www.nature.com/articles/s41...
Or the readcube link below:
rdcu.be/d5gAe
Finally we wish everyone a happy new year!!! 14/end
www.nature.com/articles/s41...
Or the readcube link below:
rdcu.be/d5gAe
Finally we wish everyone a happy new year!!! 14/end
Finally, a huge thank you to all our funders without which our work would not be possible @crick.ac.uk, Genomics England, @cancerresearchuk.org, @uclh.bsky.social @bcrfcure.bsky.social, Danish Cancer Society, UCL Cancer Institute and @cruk-cityoflondon.bsky.social. 13/14
January 3, 2025 at 11:24 AM
Finally, a huge thank you to all our funders without which our work would not be possible @crick.ac.uk, Genomics England, @cancerresearchuk.org, @uclh.bsky.social @bcrfcure.bsky.social, Danish Cancer Society, UCL Cancer Institute and @cruk-cityoflondon.bsky.social. 13/14
We thank all our collaborators @mcclellandlab.bsky.social @taponlab.bsky.social @kfugger.bsky.social @nicky Mcgranahan, Nicholas Birbak, Kevin Litchfield and Mariam Jamal-Hanjani. Thank you to the patients and their families, without which we would not be able to start this project. 12/14
January 3, 2025 at 11:24 AM
We thank all our collaborators @mcclellandlab.bsky.social @taponlab.bsky.social @kfugger.bsky.social @nicky Mcgranahan, Nicholas Birbak, Kevin Litchfield and Mariam Jamal-Hanjani. Thank you to the patients and their families, without which we would not be able to start this project. 12/14
This would not be possible w/o the bioinformatics input of @jrmblack.bsky.social @oriolpich.bsky.social Chris Bailey @ruizc.bsky.social, Paco Gimeno-Valiente, Ieva Usaite, Kerstin Thol, Maise Al Bakir & Dhruva Biswas who are dedicated to integrating real-life data with experimental results. 11/14
January 3, 2025 at 11:24 AM
This would not be possible w/o the bioinformatics input of @jrmblack.bsky.social @oriolpich.bsky.social Chris Bailey @ruizc.bsky.social, Paco Gimeno-Valiente, Ieva Usaite, Kerstin Thol, Maise Al Bakir & Dhruva Biswas who are dedicated to integrating real-life data with experimental results. 11/14
This work is co-led by @weitinglu.bsky.social and @nnennayakanu.bsky.social lab, with significant input from our long-term collaborator Jiri Bartek . @panoszalmas.bsky.social and high throughput screening core @crick.ac.uk were instrumental in starting this project. 10/14
January 3, 2025 at 11:24 AM
This work is co-led by @weitinglu.bsky.social and @nnennayakanu.bsky.social lab, with significant input from our long-term collaborator Jiri Bartek . @panoszalmas.bsky.social and high throughput screening core @crick.ac.uk were instrumental in starting this project. 10/14
Now, which of the FAT1 KO phenotypes are due to YAP1 localization? By depleting YAP1 in FAT1 KO cells, we could fully rescue the cytokinesis failure in FAT1 KO. cells, suggesting this is YAP1 dependent. However, the ⏫mitotic error rate was not dependent on YAP1 dysregulation 9/14
January 3, 2025 at 11:24 AM
Now, which of the FAT1 KO phenotypes are due to YAP1 localization? By depleting YAP1 in FAT1 KO cells, we could fully rescue the cytokinesis failure in FAT1 KO. cells, suggesting this is YAP1 dependent. However, the ⏫mitotic error rate was not dependent on YAP1 dysregulation 9/14
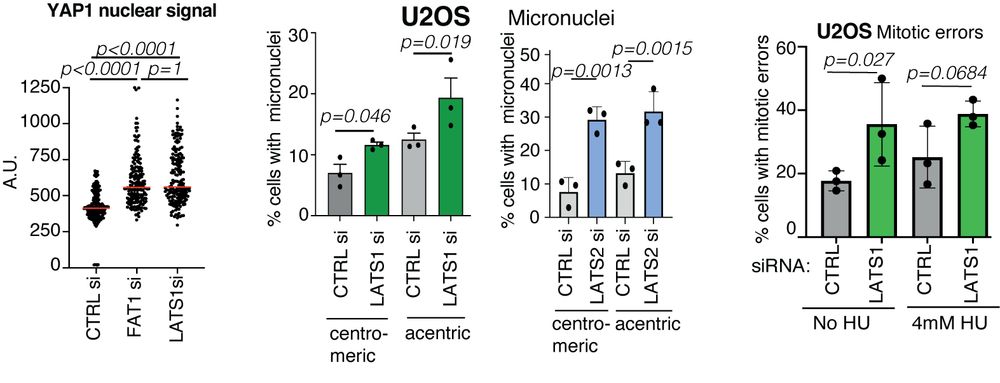
How does FAT1 loss lead to its phenotype? We noticed that similar to another key member of the Hippo🦛 pathway LATS1, FAT1 depletion enhances YAP1 nuclear signal. Perhaps LATS1/2 loss contribute to the DDR defects and the phenotypes might be somehow dependent on YAP1? 8/14
January 3, 2025 at 11:24 AM
How does FAT1 loss lead to its phenotype? We noticed that similar to another key member of the Hippo🦛 pathway LATS1, FAT1 depletion enhances YAP1 nuclear signal. Perhaps LATS1/2 loss contribute to the DDR defects and the phenotypes might be somehow dependent on YAP1? 8/14
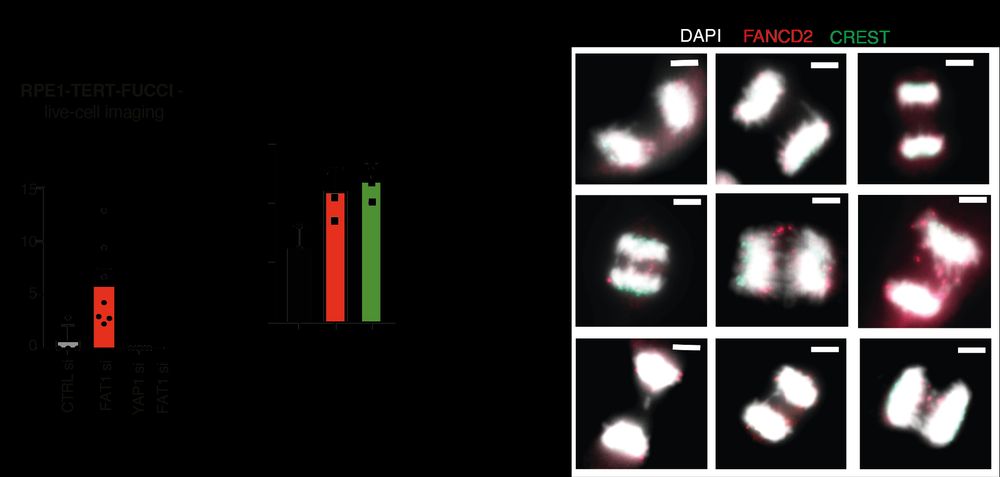
How does FAT1 loss cause WGD? Using the FUCCI system in RPE1 cells (thanks to John Diffley lab @crick.ac.uk ) we could track the fidelity of cell division in real-time. The cell without FAT1 failed cytokinesis, could not finish mitosis and remained as 1 daughter cell with 2x the genome content 7/14
January 3, 2025 at 11:24 AM
How does FAT1 loss cause WGD? Using the FUCCI system in RPE1 cells (thanks to John Diffley lab @crick.ac.uk ) we could track the fidelity of cell division in real-time. The cell without FAT1 failed cytokinesis, could not finish mitosis and remained as 1 daughter cell with 2x the genome content 7/14
We also used live cell imaging to quantify mitotic errors. 6/14
January 3, 2025 at 11:24 AM
We also used live cell imaging to quantify mitotic errors. 6/14
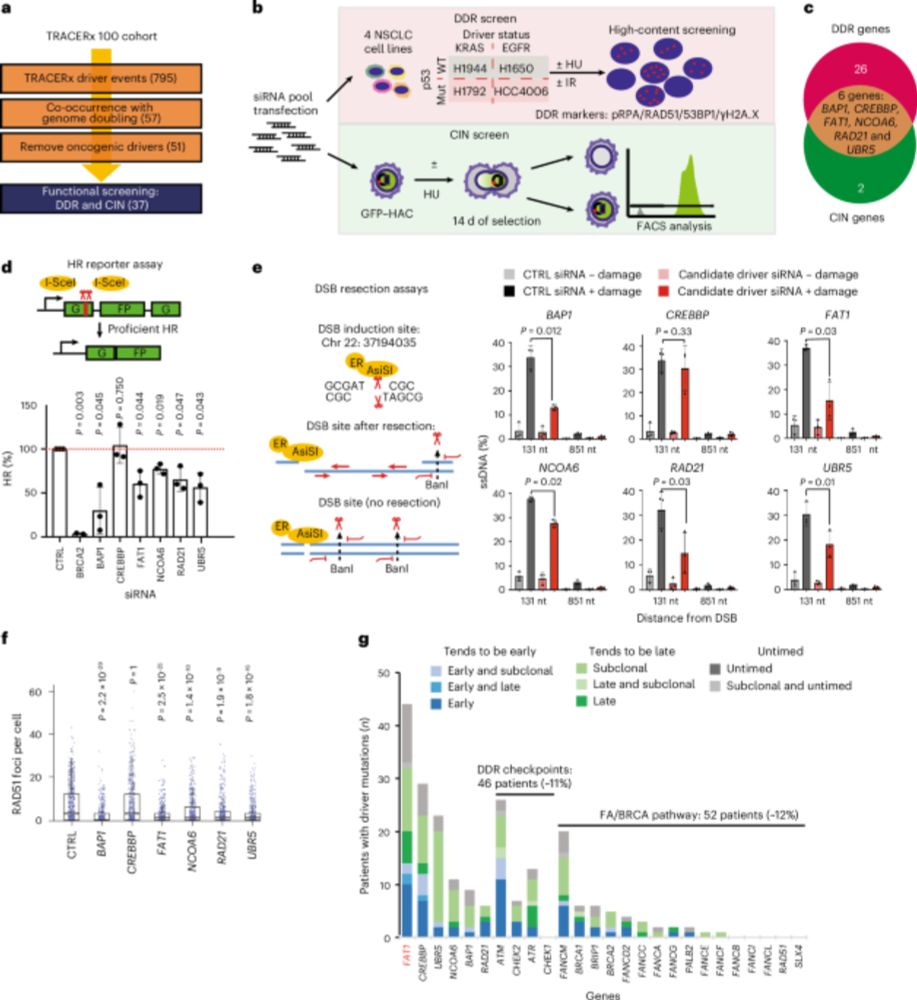
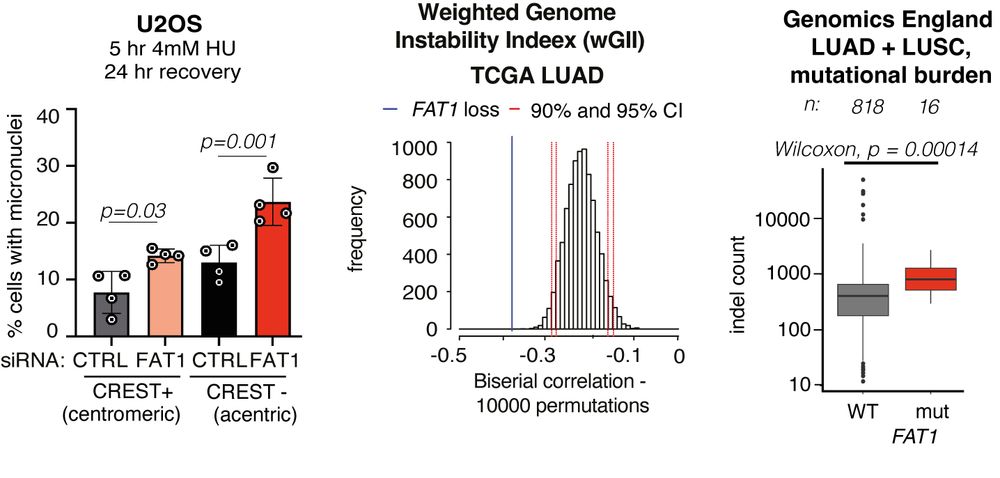
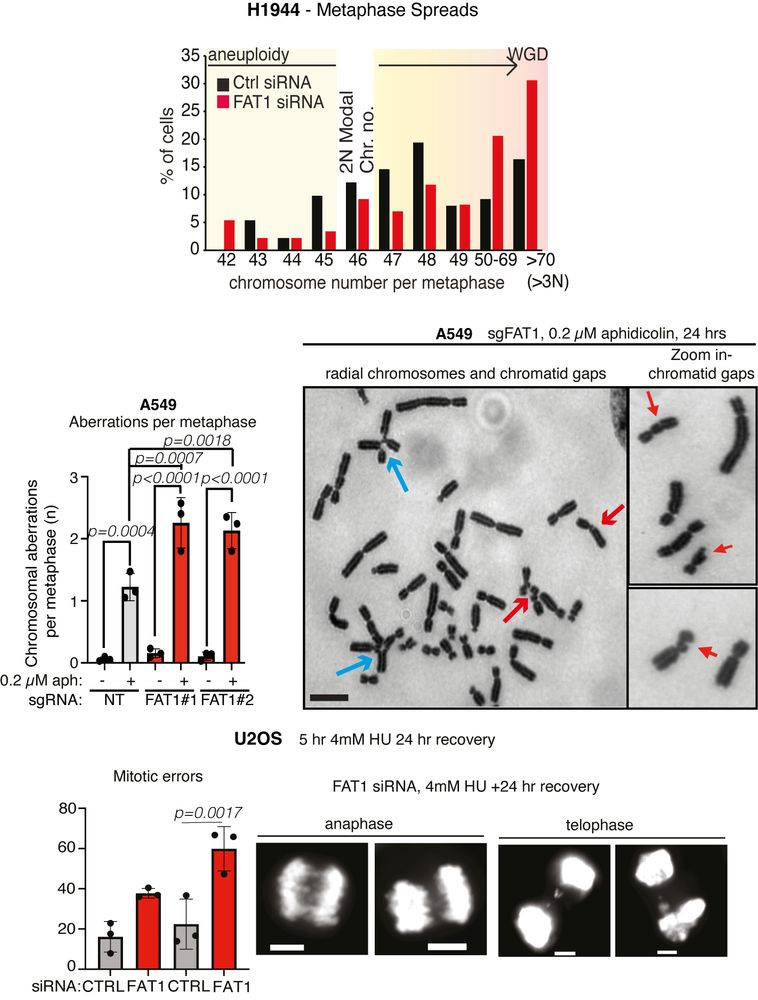
Focusing on CIN, FAT1 loss increased micronuclei formation. FAT1mutant tumours exhibited a higher mutational burden. FAT1 depletion increased ⏫ chromosome number and ⏫radial chromosome structures (blue arrows) and we also observed an⏫ rate of mitotic defects. 5/14
January 3, 2025 at 11:24 AM
Focusing on CIN, FAT1 loss increased micronuclei formation. FAT1mutant tumours exhibited a higher mutational burden. FAT1 depletion increased ⏫ chromosome number and ⏫radial chromosome structures (blue arrows) and we also observed an⏫ rate of mitotic defects. 5/14
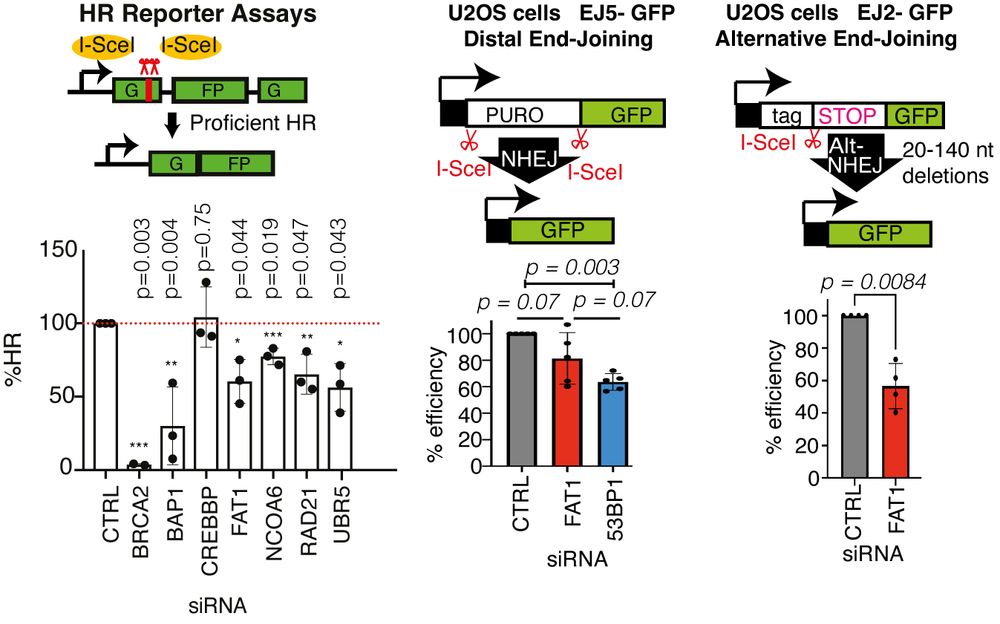
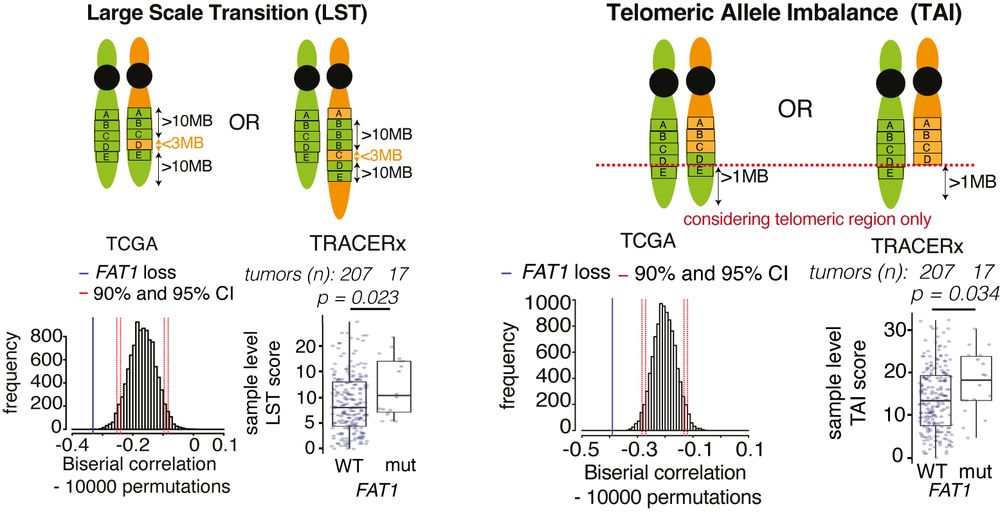
Experimentally, FAT1 depletion reduced ⏬end-resection at DNA double-strand breaks, ⏬HR repair and alternative end-joining, but did not affect cNHEJ. Using TCGA and TRACERx data, we confirmed that FAT1mutant tumours exhibited hallmarks of HR deficiencies like TAI and LST. 4/14
January 3, 2025 at 11:24 AM
Experimentally, FAT1 depletion reduced ⏬end-resection at DNA double-strand breaks, ⏬HR repair and alternative end-joining, but did not affect cNHEJ. Using TCGA and TRACERx data, we confirmed that FAT1mutant tumours exhibited hallmarks of HR deficiencies like TAI and LST. 4/14
We devised 2 screens to identify drivers that might contribute to both deficiencies in the DNA damage response (DDR) and cause CIN. We focused on a gene called FAT1 (4q35.2), which is extensively mutated/lost in NSCLC before WGD, but we knew little about its function. 3/14
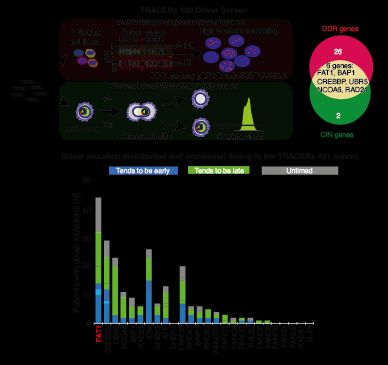
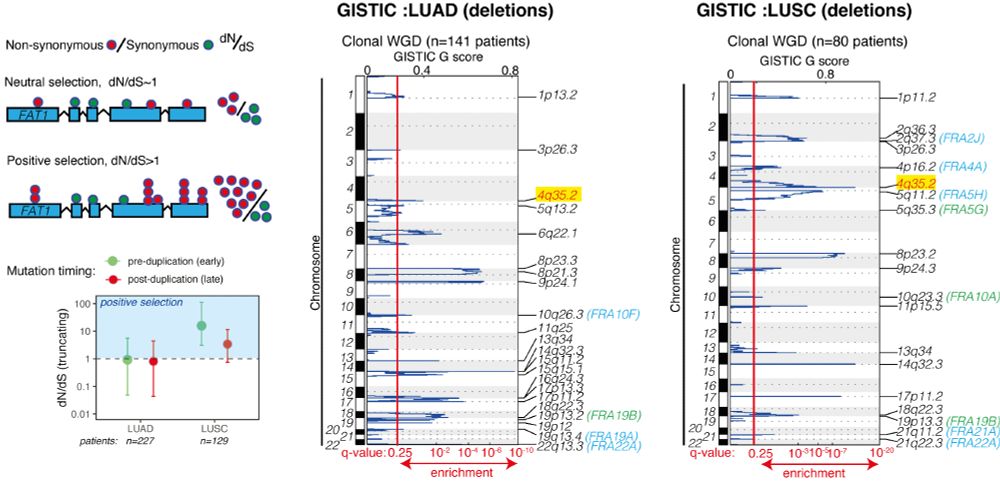
January 3, 2025 at 11:24 AM
We devised 2 screens to identify drivers that might contribute to both deficiencies in the DNA damage response (DDR) and cause CIN. We focused on a gene called FAT1 (4q35.2), which is extensively mutated/lost in NSCLC before WGD, but we knew little about its function. 3/14

