Wei-Ting Lu
@weitinglu.bsky.social
Tea addict and scientist, in that order. Incoming Group Leader, Dept of Oncology, University of Oxford. All views my own.
Now, which of the FAT1 KO phenotypes are due to YAP1 localization? By depleting YAP1 in FAT1 KO cells, we could fully rescue the cytokinesis failure in FAT1 KO. cells, suggesting this is YAP1 dependent. However, the ⏫mitotic error rate was not dependent on YAP1 dysregulation 9/14
January 3, 2025 at 11:24 AM
Now, which of the FAT1 KO phenotypes are due to YAP1 localization? By depleting YAP1 in FAT1 KO cells, we could fully rescue the cytokinesis failure in FAT1 KO. cells, suggesting this is YAP1 dependent. However, the ⏫mitotic error rate was not dependent on YAP1 dysregulation 9/14
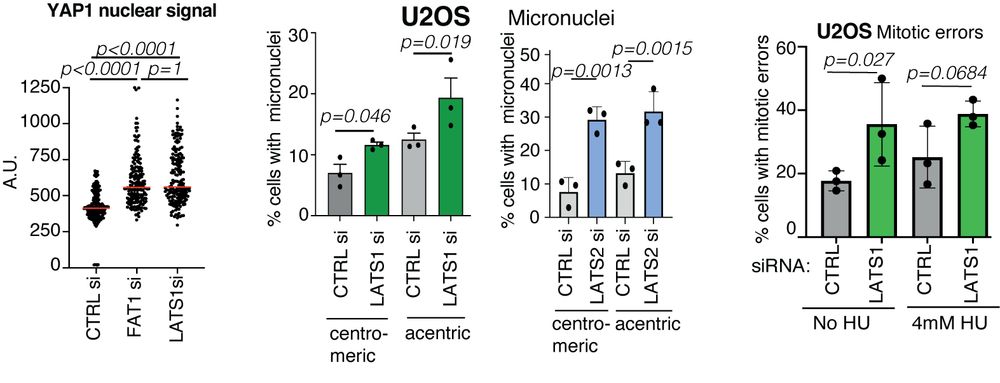
How does FAT1 loss lead to its phenotype? We noticed that similar to another key member of the Hippo🦛 pathway LATS1, FAT1 depletion enhances YAP1 nuclear signal. Perhaps LATS1/2 loss contribute to the DDR defects and the phenotypes might be somehow dependent on YAP1? 8/14
January 3, 2025 at 11:24 AM
How does FAT1 loss lead to its phenotype? We noticed that similar to another key member of the Hippo🦛 pathway LATS1, FAT1 depletion enhances YAP1 nuclear signal. Perhaps LATS1/2 loss contribute to the DDR defects and the phenotypes might be somehow dependent on YAP1? 8/14
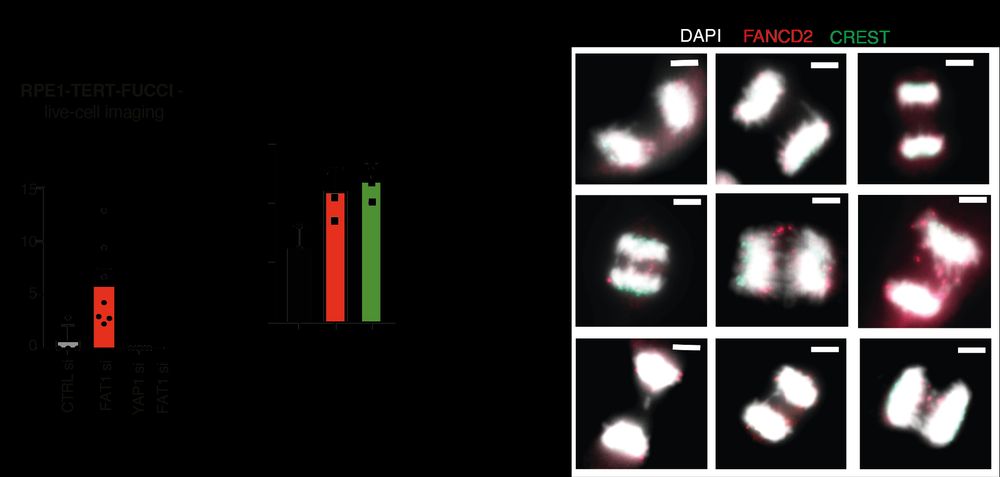
How does FAT1 loss cause WGD? Using the FUCCI system in RPE1 cells (thanks to John Diffley lab @crick.ac.uk ) we could track the fidelity of cell division in real-time. The cell without FAT1 failed cytokinesis, could not finish mitosis and remained as 1 daughter cell with 2x the genome content 7/14
January 3, 2025 at 11:24 AM
How does FAT1 loss cause WGD? Using the FUCCI system in RPE1 cells (thanks to John Diffley lab @crick.ac.uk ) we could track the fidelity of cell division in real-time. The cell without FAT1 failed cytokinesis, could not finish mitosis and remained as 1 daughter cell with 2x the genome content 7/14
We also used live cell imaging to quantify mitotic errors. 6/14
January 3, 2025 at 11:24 AM
We also used live cell imaging to quantify mitotic errors. 6/14
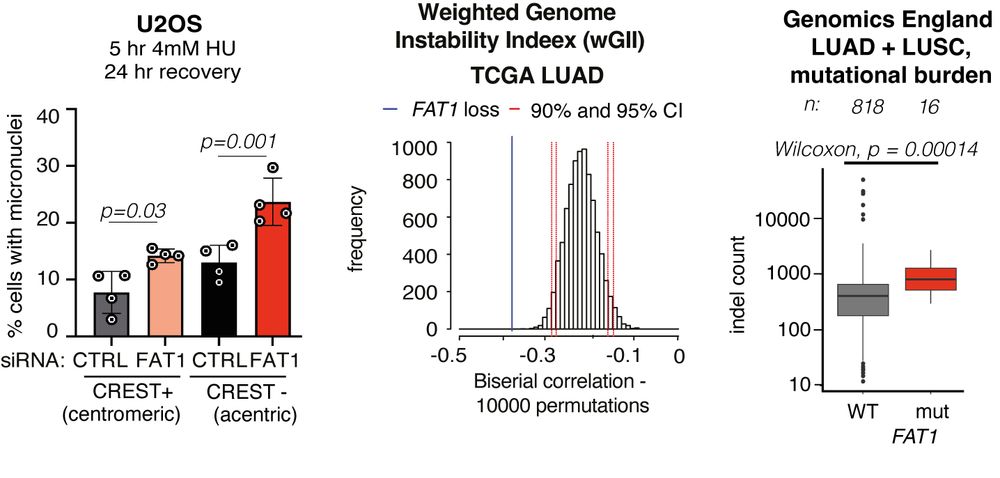
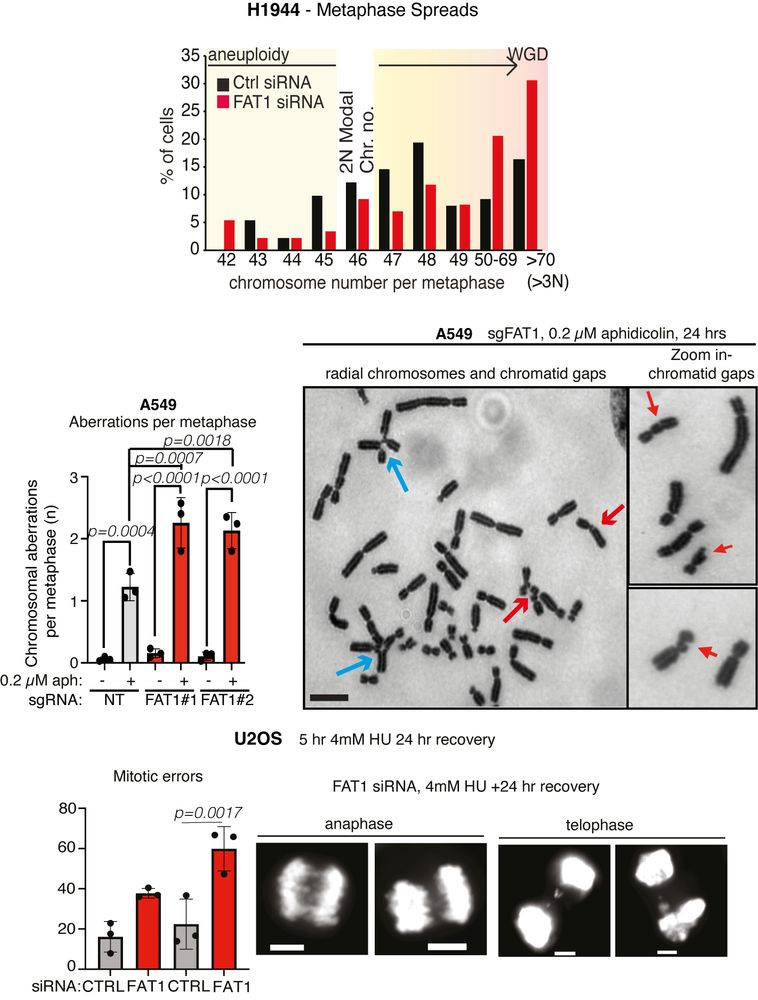
Focusing on CIN, FAT1 loss increased micronuclei formation. FAT1mutant tumours exhibited a higher mutational burden. FAT1 depletion increased ⏫ chromosome number and ⏫radial chromosome structures (blue arrows) and we also observed an⏫ rate of mitotic defects. 5/14
January 3, 2025 at 11:24 AM
Focusing on CIN, FAT1 loss increased micronuclei formation. FAT1mutant tumours exhibited a higher mutational burden. FAT1 depletion increased ⏫ chromosome number and ⏫radial chromosome structures (blue arrows) and we also observed an⏫ rate of mitotic defects. 5/14
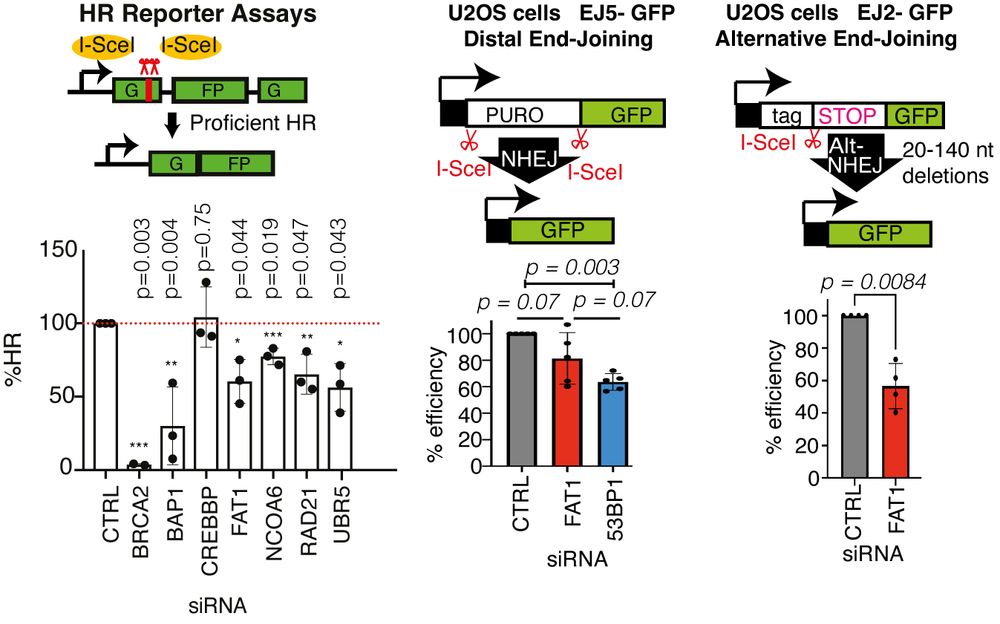
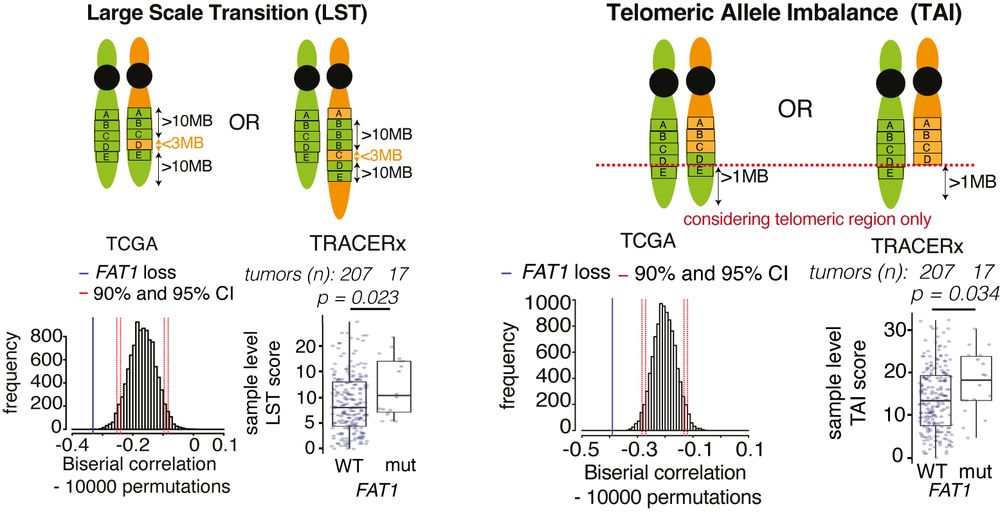
Experimentally, FAT1 depletion reduced ⏬end-resection at DNA double-strand breaks, ⏬HR repair and alternative end-joining, but did not affect cNHEJ. Using TCGA and TRACERx data, we confirmed that FAT1mutant tumours exhibited hallmarks of HR deficiencies like TAI and LST. 4/14
January 3, 2025 at 11:24 AM
Experimentally, FAT1 depletion reduced ⏬end-resection at DNA double-strand breaks, ⏬HR repair and alternative end-joining, but did not affect cNHEJ. Using TCGA and TRACERx data, we confirmed that FAT1mutant tumours exhibited hallmarks of HR deficiencies like TAI and LST. 4/14
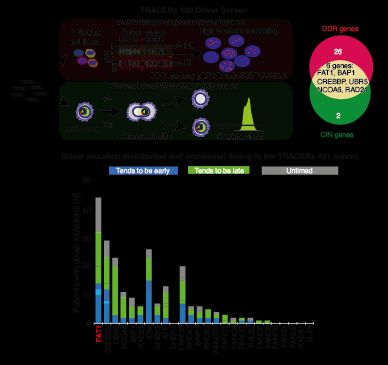
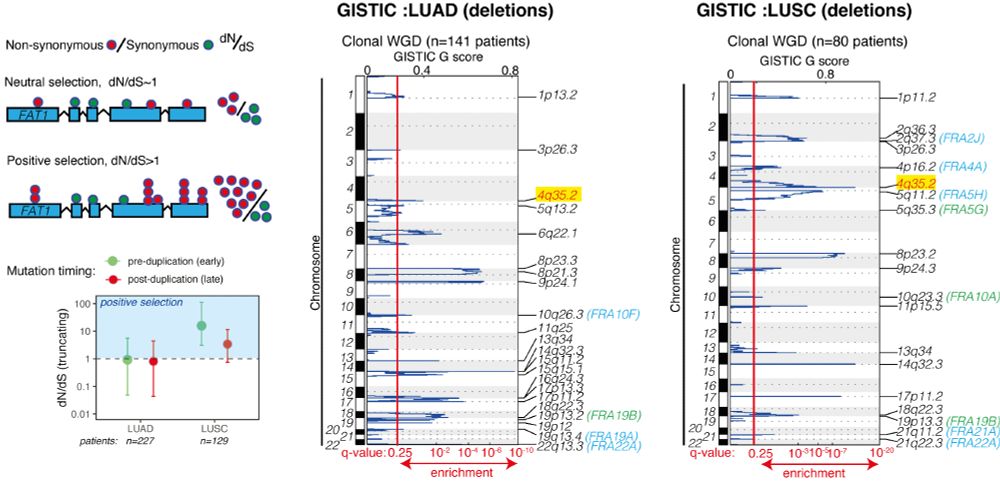
We devised 2 screens to identify drivers that might contribute to both deficiencies in the DNA damage response (DDR) and cause CIN. We focused on a gene called FAT1 (4q35.2), which is extensively mutated/lost in NSCLC before WGD, but we knew little about its function. 3/14
January 3, 2025 at 11:24 AM
We devised 2 screens to identify drivers that might contribute to both deficiencies in the DNA damage response (DDR) and cause CIN. We focused on a gene called FAT1 (4q35.2), which is extensively mutated/lost in NSCLC before WGD, but we knew little about its function. 3/14
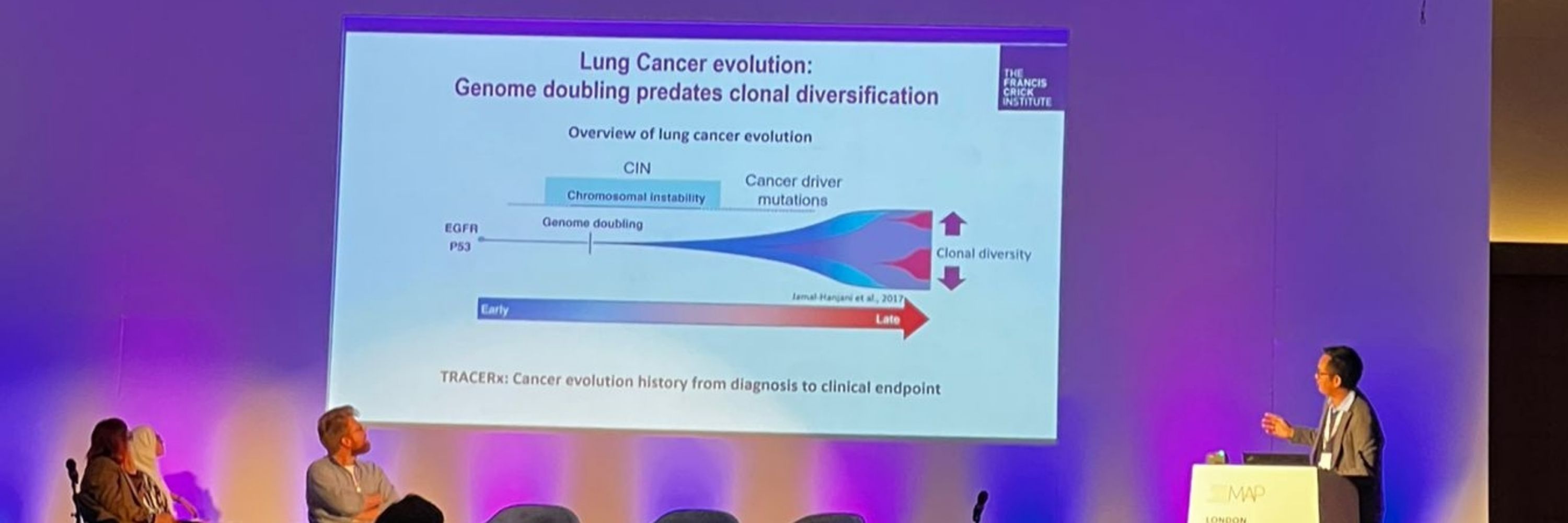
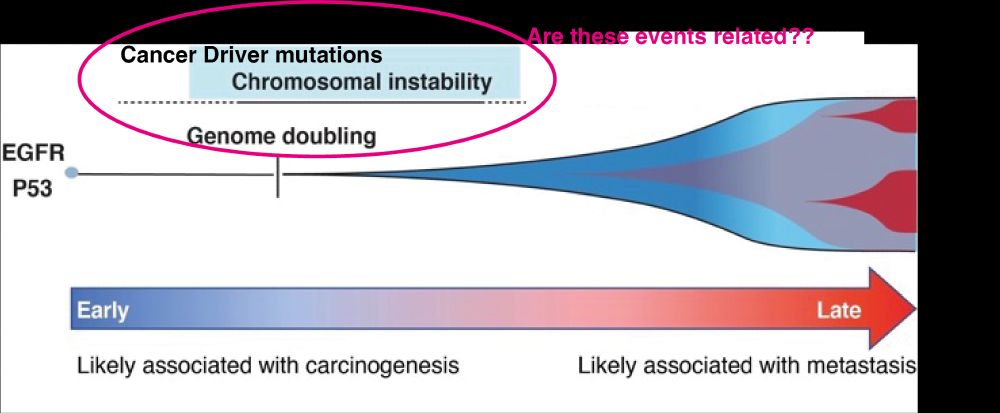
This journey started 7 years ago when we explored the cancer evolution data from TRACERx100, and observed that clonal diversification cooccurs with whole genome doubling (WGD) & extensive chromosomal instability (CIN). Might this be triggered by specific cancer driver events? 2/14
January 3, 2025 at 11:24 AM
This journey started 7 years ago when we explored the cancer evolution data from TRACERx100, and observed that clonal diversification cooccurs with whole genome doubling (WGD) & extensive chromosomal instability (CIN). Might this be triggered by specific cancer driver events? 2/14

