🔬 Through Vizgen Lab Services, Marco Genua (Fondazione Telethon) mapped the tumor microenvironment, overcoming RNA quality challenges.
🔗 hubs.ly/Q038VZgB0

🔬 Through Vizgen Lab Services, Marco Genua (Fondazione Telethon) mapped the tumor microenvironment, overcoming RNA quality challenges.
🔗 hubs.ly/Q038VZgB0
Read the full pre-print: t.co/RmW36N6zni
#SpatialBiology #MERSCOPE #MERFISH

Read the full pre-print: t.co/RmW36N6zni
#SpatialBiology #MERSCOPE #MERFISH
Read here: www.cell.com/cell-reports...

Read here: www.cell.com/cell-reports...
Learn more in today’s announcement: vizgen.com/vizgen-expan...

Learn more in today’s announcement: vizgen.com/vizgen-expan...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
We're excited to share our four Keynote Speakers discussing the latest research, technologies, and applications across the field.
Register ➡️ https://buff.ly/4ifjTib

We're excited to share our four Keynote Speakers discussing the latest research, technologies, and applications across the field.
Register ➡️ https://buff.ly/4ifjTib
Research using #MERFISH reveals stable, long-range chromosome loops over 100 Mb, driven by nucleation centers. This offers new understanding of chromatin organization and cellular function.
#Genomics #ChromosomeFolding #MERFISH
www.biorxiv.org/content/10.1...

Research using #MERFISH reveals stable, long-range chromosome loops over 100 Mb, driven by nucleation centers. This offers new understanding of chromatin organization and cellular function.
#Genomics #ChromosomeFolding #MERFISH
www.biorxiv.org/content/10.1...
#MERFISH #BacterialTranscriptomics #Microbiology
www.science.org/doi/10.1126/...

#MERFISH #BacterialTranscriptomics #Microbiology
www.science.org/doi/10.1126/...
#MERFISH #MERSCOPE
www.biorxiv.org/content/10.1...

#MERFISH #MERSCOPE
www.biorxiv.org/content/10.1...
#MERFISH #SpatialTranscriptomics #SpatialBiology #SingleCell #SubCellular #OnlyRealDotsCount

#MERFISH #SpatialTranscriptomics #SpatialBiology #SingleCell #SubCellular #OnlyRealDotsCount
#MERFISH #BacterialTranscriptomics #Microbiology
www.science.org/doi/10.1126/...

#MERFISH #BacterialTranscriptomics #Microbiology
www.science.org/doi/10.1126/...
Research using #MERFISH reveals stable, long-range chromosome loops over 100 Mb, driven by nucleation centers. This offers new understanding of chromatin organization and cellular function.
#Genomics #ChromosomeFolding #MERFISH
www.biorxiv.org/content/10.1...

Research using #MERFISH reveals stable, long-range chromosome loops over 100 Mb, driven by nucleation centers. This offers new understanding of chromatin organization and cellular function.
#Genomics #ChromosomeFolding #MERFISH
www.biorxiv.org/content/10.1...

“Bacterial-MERFISH” provides ~1000-fold volumetric expansion of individual cells, charts gene expression in hundreds of thousands of cells, deciphering bacterial single-cell heterogeneity, intracellular transcriptome organization, and bacterial adaptation to µm-scale niches in vivo
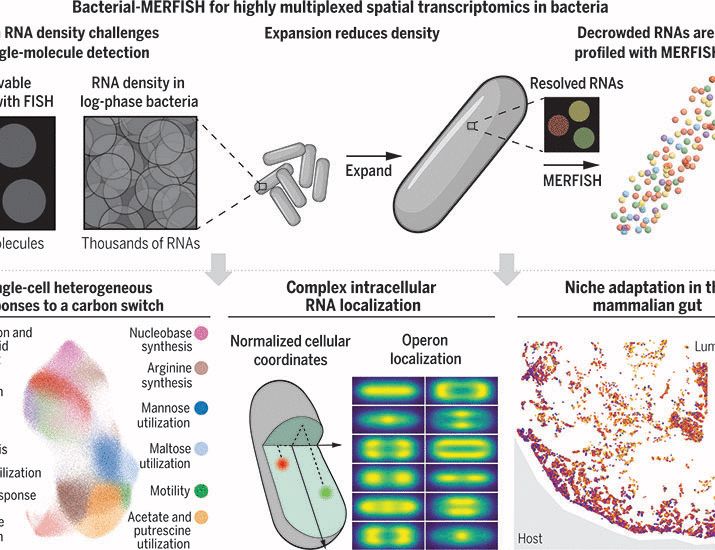
“Bacterial-MERFISH” provides ~1000-fold volumetric expansion of individual cells, charts gene expression in hundreds of thousands of cells, deciphering bacterial single-cell heterogeneity, intracellular transcriptome organization, and bacterial adaptation to µm-scale niches in vivo
Abstract submission is open for PhD and Early-Career researchers to present a short talk or poster during the symposium.
buff.ly/3WPzXyJ

Abstract submission is open for PhD and Early-Career researchers to present a short talk or poster during the symposium.
buff.ly/3WPzXyJ
Thanks to #MERFISH spatial transcriptomics a team uncovered how tissue origin and vascular positioning shape blood vessel formation.
#SpatialTranscriptomics #MERFISH #SingleCell #VascularDevelopment #CRISPR
www.ahajournals.org/doi/10.1161/...

Thanks to #MERFISH spatial transcriptomics a team uncovered how tissue origin and vascular positioning shape blood vessel formation.
#SpatialTranscriptomics #MERFISH #SingleCell #VascularDevelopment #CRISPR
www.ahajournals.org/doi/10.1161/...
#MERFISH #SpatialTranscriptomics #SpatialBiology #SingleCell #SubCellular #OnlyRealDotsCount

#MERFISH #SpatialTranscriptomics #SpatialBiology #SingleCell #SubCellular #OnlyRealDotsCount
#SpatialTranscriptomics @whatchamacaulay.bsky.social

#SpatialTranscriptomics @whatchamacaulay.bsky.social
A great collab with @katielong.bsky.social and @cristobaluauy.bsky.social at JIC, and Ashleigh Lister and the #SingleCell team at Earlham Institute.
A great collab with @katielong.bsky.social and @cristobaluauy.bsky.social at JIC, and Ashleigh Lister and the #SingleCell team at Earlham Institute.

We're excited to share our four Keynote Speakers discussing the latest research, technologies, and applications across the field.
Register ➡️ www.earlham.ac.uk/events/norwi...

We're excited to share our four Keynote Speakers discussing the latest research, technologies, and applications across the field.
Register ➡️ www.earlham.ac.uk/events/norwi...
@johninnescentre.bsky.social & @earlhaminst.bsky.social!
Using #MERFISH, the team resolved the expression of 200 genes to cellular resolution, uncovering remarkable insights into inflorescence development.
www.biorxiv.org/content/10.1...
@johninnescentre.bsky.social & @earlhaminst.bsky.social!
Using #MERFISH, the team resolved the expression of 200 genes to cellular resolution, uncovering remarkable insights into inflorescence development.
www.biorxiv.org/content/10.1...
#Spatial #Genomics

#Spatial #Genomics



