puheenvuoro.uusisuomi.fi/villekalervo...

puheenvuoro.uusisuomi.fi/villekalervo...

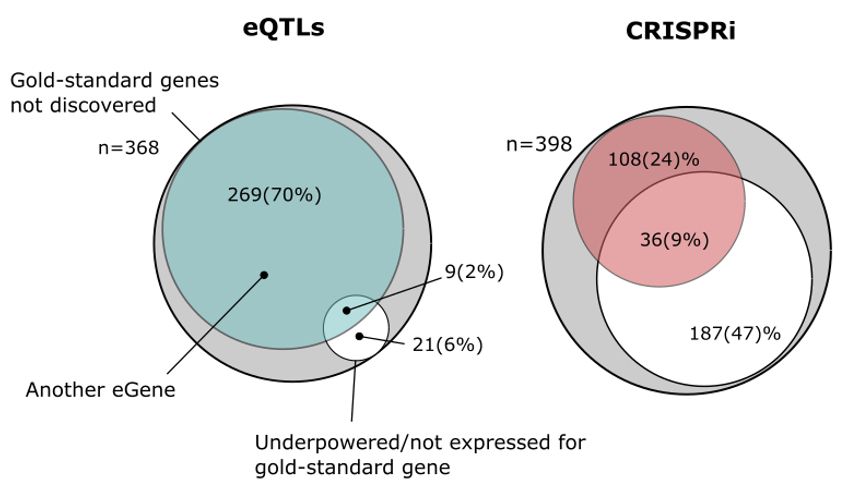
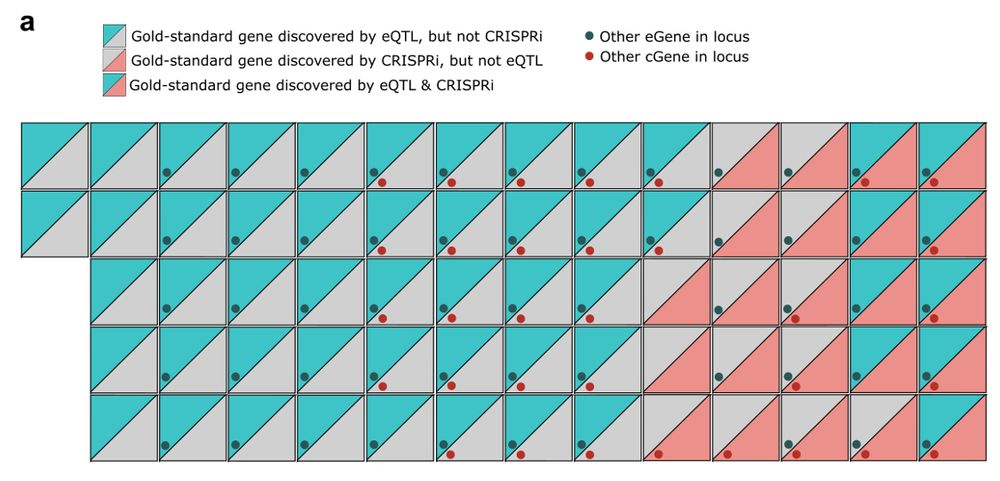

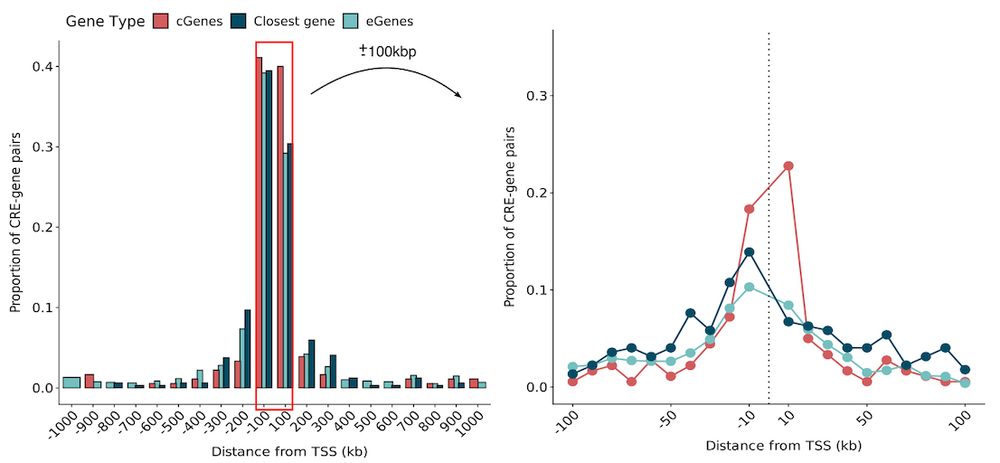
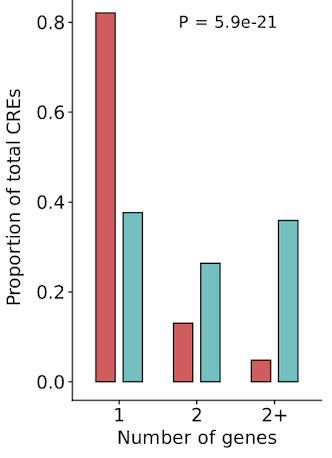
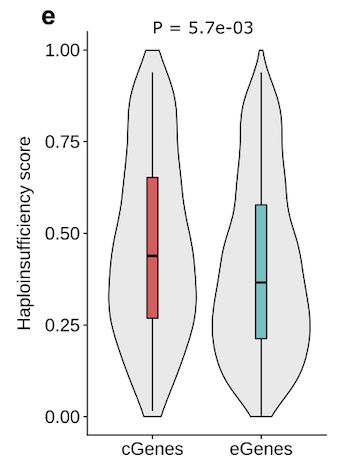
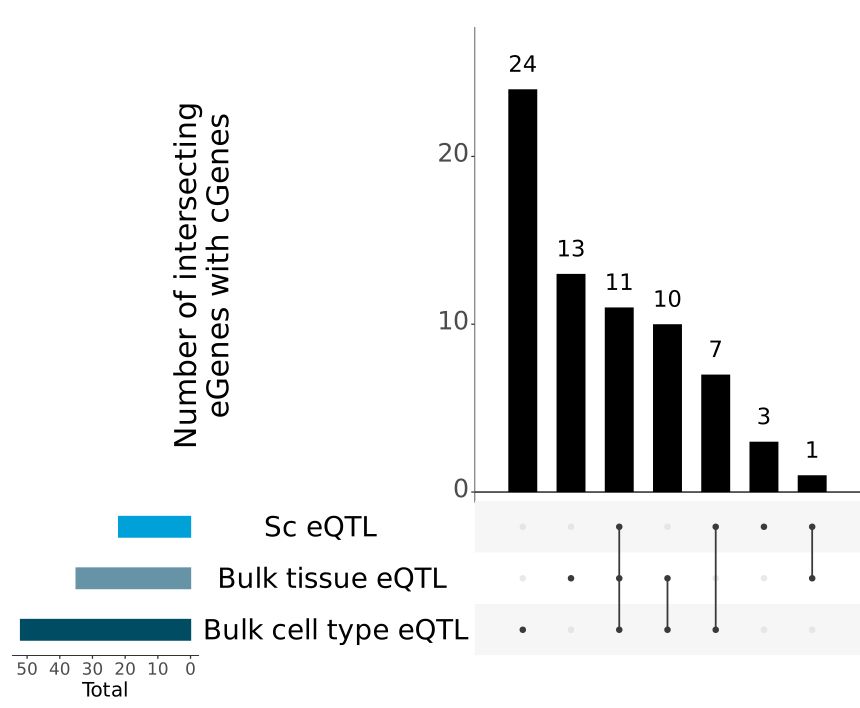


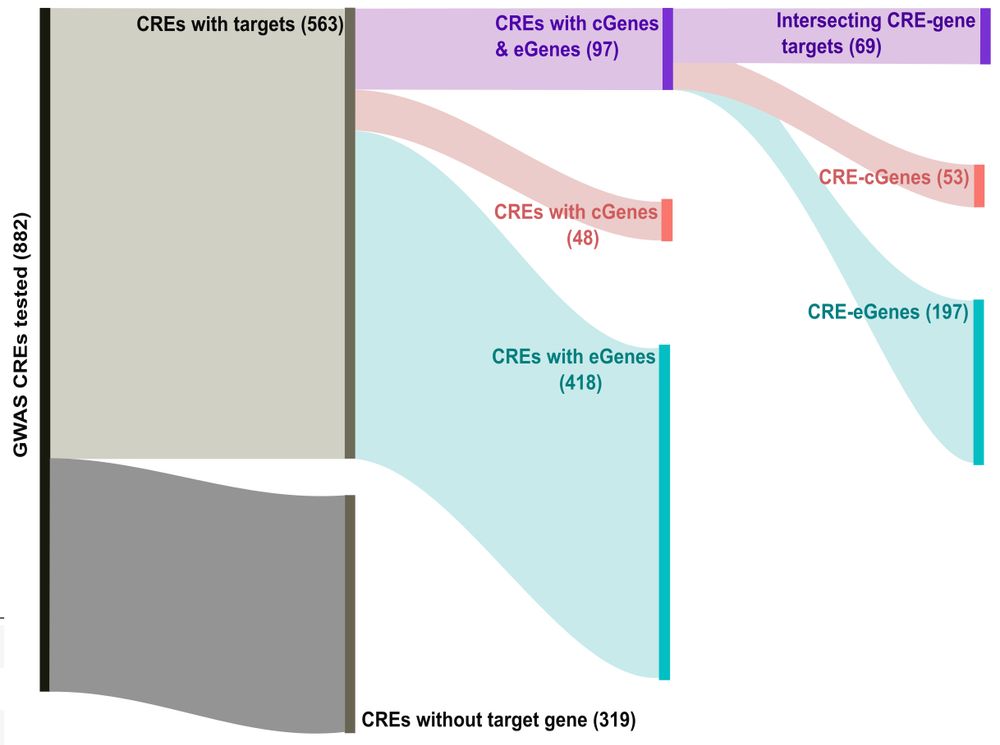
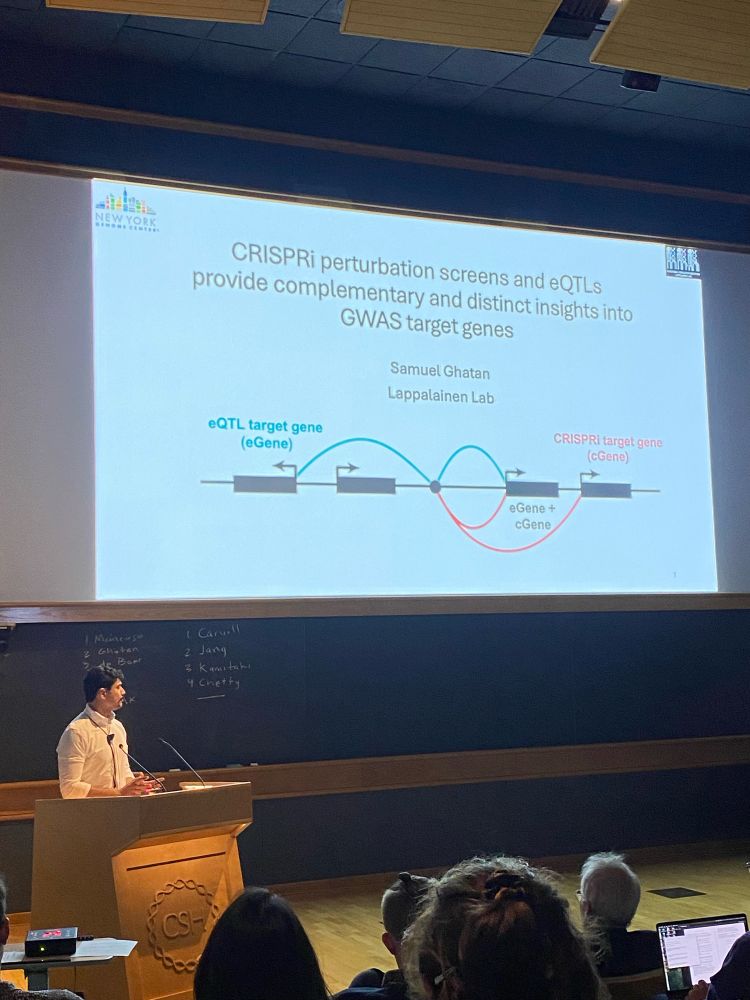
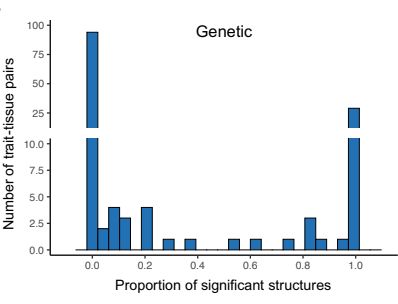
www.science.org/doi/10.1126/...
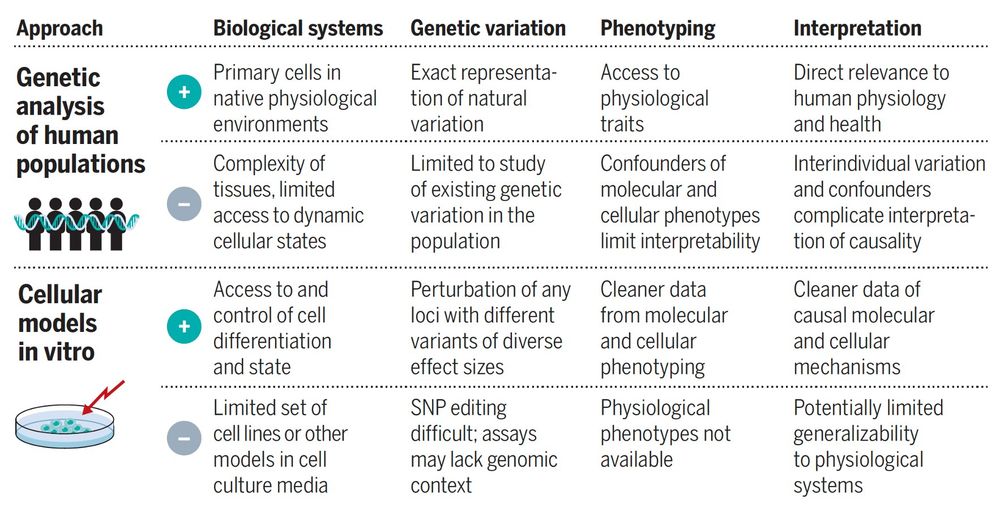
www.science.org/doi/10.1126/...
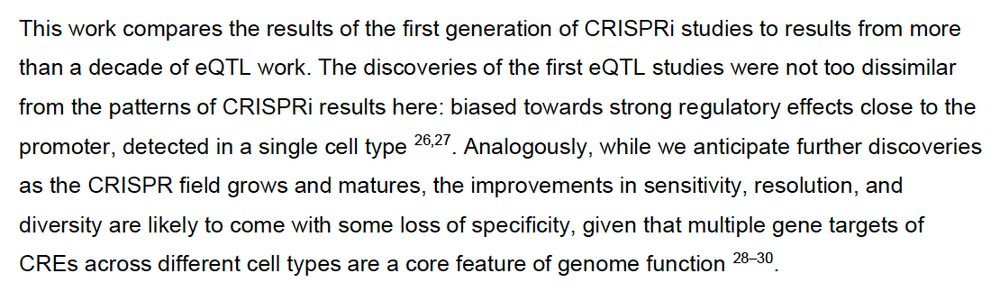
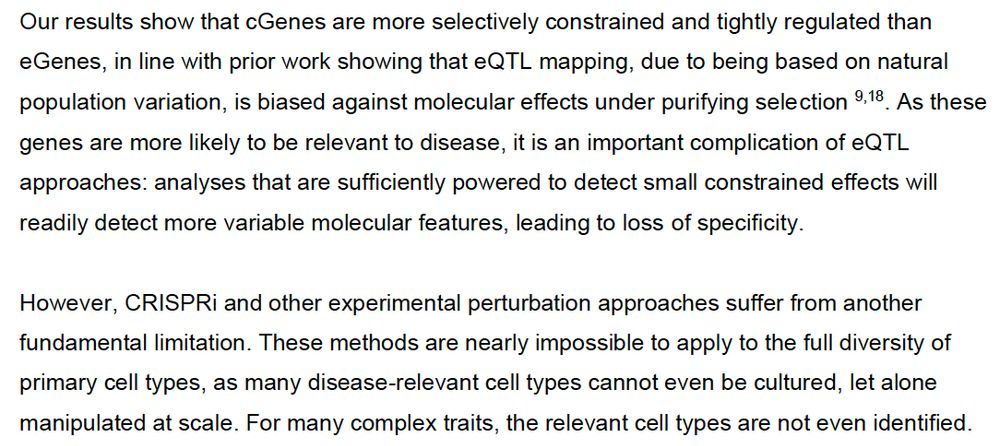
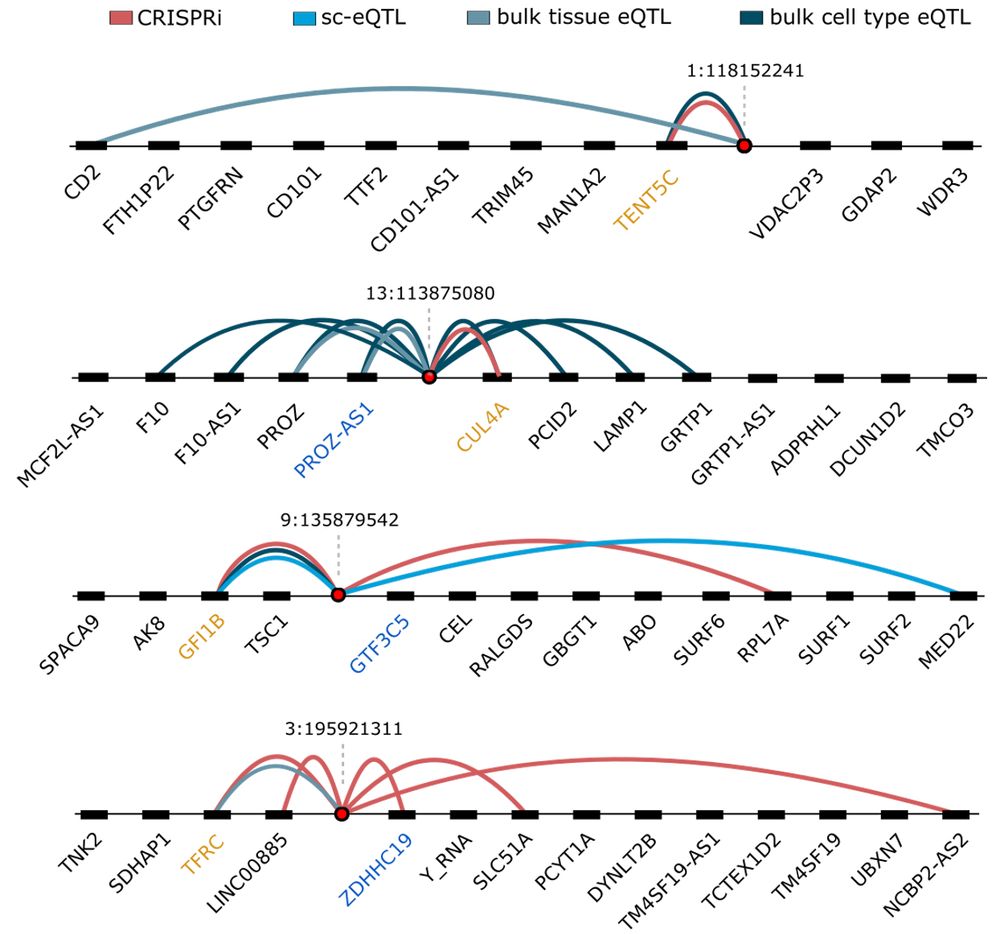
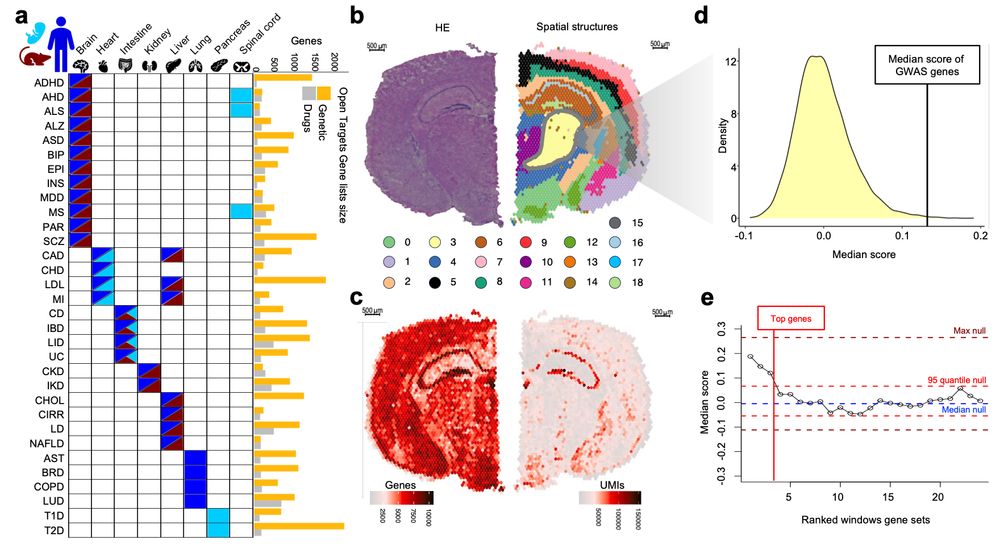
www.biorxiv.org/content/10.1...
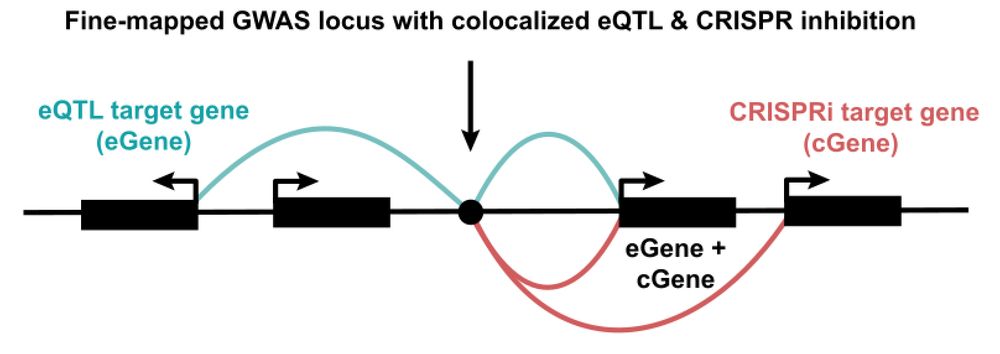
www.biorxiv.org/content/10.1...
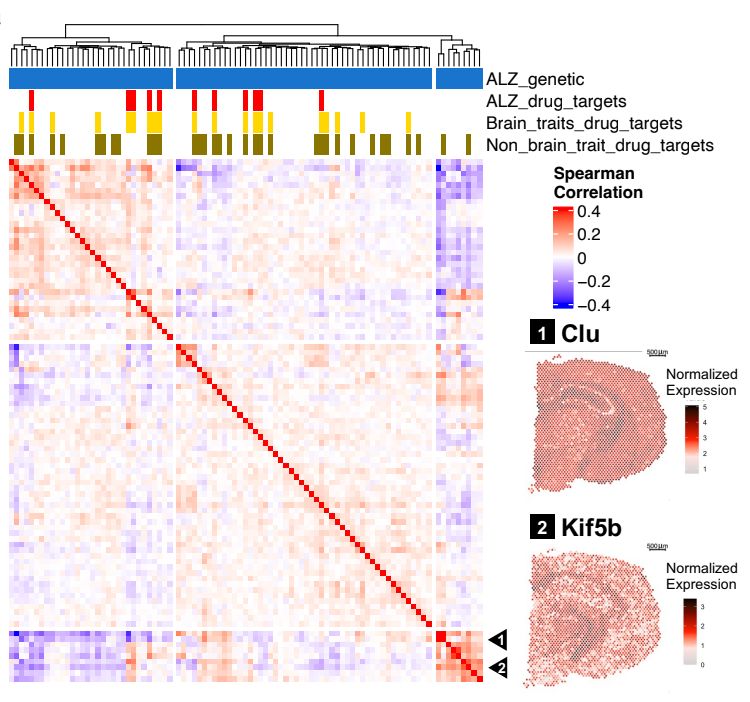
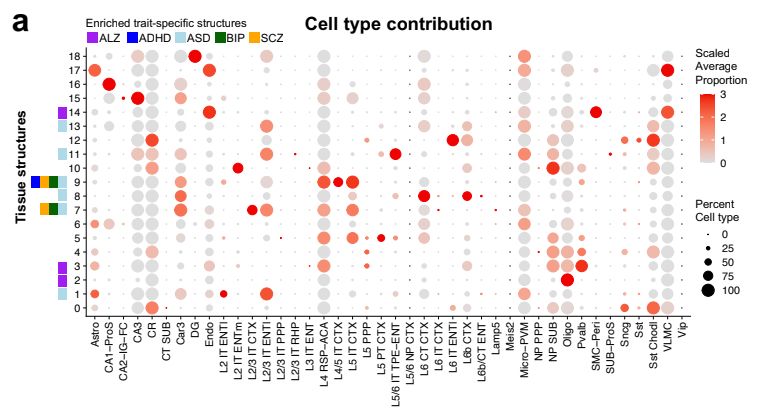
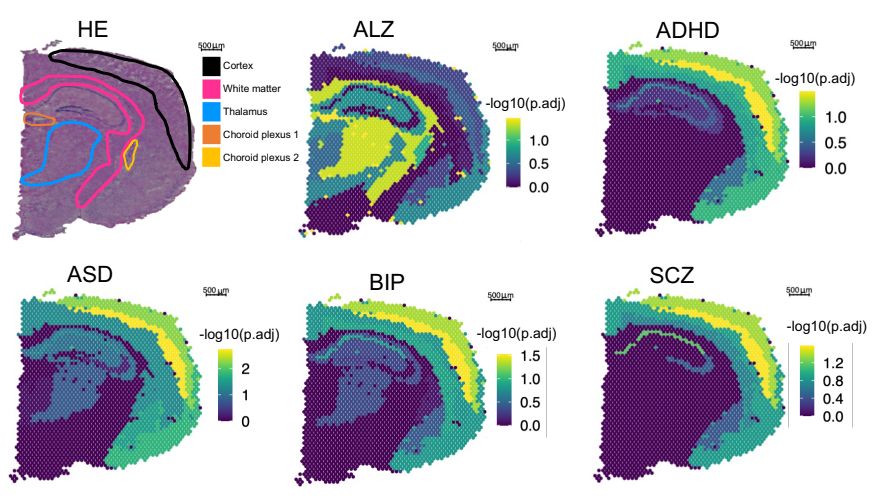




We're excited to bring PhD students together with an all-star list of leaders in human genetics.
📅 July 27-31, 2025
📍 Wellcome Genome Campus, UK
📝 Apply by March 7 at www.lpshg.com

We're excited to bring PhD students together with an all-star list of leaders in human genetics.
📅 July 27-31, 2025
📍 Wellcome Genome Campus, UK
📝 Apply by March 7 at www.lpshg.com