Thea Schulze (Lindorff-Larsen Lab): Predicting mutated protein abundance @tkschulze.bsky.social
Taylor Mighell (Lehner Lab): Massive mutagenesis to understand GPCRs @taylor-mighell.bsky.social
🔗 More info at varianteffect.org/seminar-series
@varianteffect.bsky.social

Thea Schulze (Lindorff-Larsen Lab): Predicting mutated protein abundance @tkschulze.bsky.social
Taylor Mighell (Lehner Lab): Massive mutagenesis to understand GPCRs @taylor-mighell.bsky.social
🔗 More info at varianteffect.org/seminar-series
@varianteffect.bsky.social
See preprint for:
— Ensembles of >12000 full-length human proteins
— Analysis of IDRs in >1500 TFs
📜 doi.org/10.1101/2025...
💾 github.com/KULL-Centre/...

See preprint for:
— Ensembles of >12000 full-length human proteins
— Analysis of IDRs in >1500 TFs
📜 doi.org/10.1101/2025...
💾 github.com/KULL-Centre/...
A coarse-grained model for disordered proteins under crowded conditions
(that is the CALVADOS PEG model) is now published in final form:
dx.doi.org/10.1002/pro....
@asrauh.bsky.social @giuliotesei.bsky.social

A coarse-grained model for disordered proteins under crowded conditions
(that is the CALVADOS PEG model) is now published in final form:
dx.doi.org/10.1002/pro....
@asrauh.bsky.social @giuliotesei.bsky.social
📜 www.biorxiv.org/content/10.1...
🖥️ github.com/KULL-Centre/...

📜 www.biorxiv.org/content/10.1...
🖥️ github.com/KULL-Centre/...
Big thanks to @sobuelow.bsky.social, @lindorfflarsen.bsky.social, and the whole team for making this possible.
Thrilled to mark this as my first last-author paper!
We’ve got your back!
@sobuelow.bsky.social & @giuliotesei.bsky.social—together with the rest of the team—describe our software for simulations using the CALVADOS models incl. recipes for several applications. 1/5
doi.org/10.48550/arX...
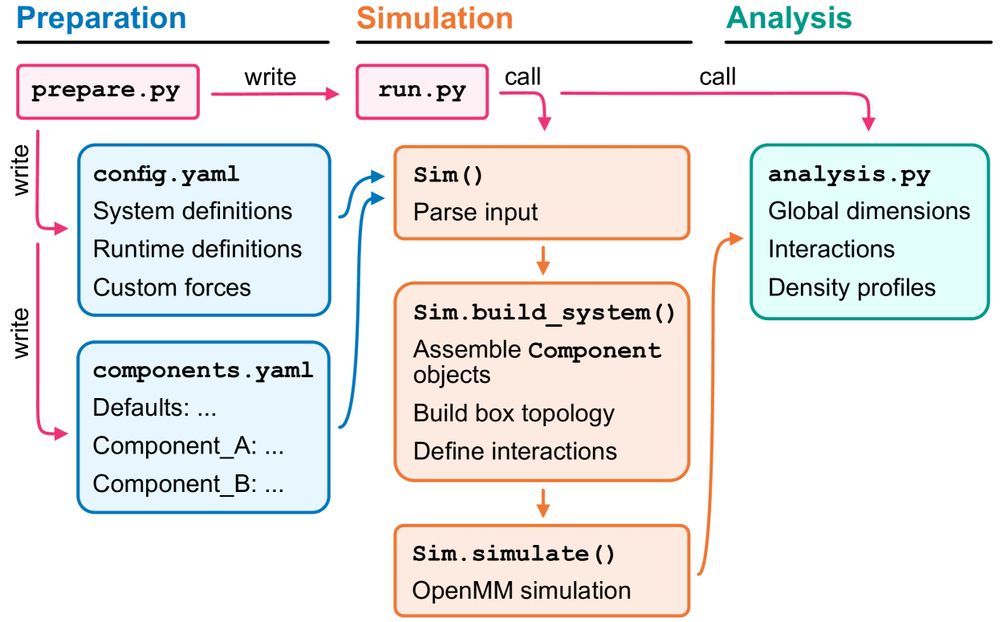
Big thanks to @sobuelow.bsky.social, @lindorfflarsen.bsky.social, and the whole team for making this possible.
Thrilled to mark this as my first last-author paper!
We’ve got your back!
@sobuelow.bsky.social & @giuliotesei.bsky.social—together with the rest of the team—describe our software for simulations using the CALVADOS models incl. recipes for several applications. 1/5
doi.org/10.48550/arX...

We’ve got your back!
@sobuelow.bsky.social & @giuliotesei.bsky.social—together with the rest of the team—describe our software for simulations using the CALVADOS models incl. recipes for several applications. 1/5
doi.org/10.48550/arX...
Here @tkschulze.bsky.social devised a strategy to take this into account while training models
www.biorxiv.org/content/10.1...

Here @tkschulze.bsky.social devised a strategy to take this into account while training models
www.biorxiv.org/content/10.1...
We find that much of the variation can be explained and predicted by a burial-dependent substitution matrix
Lots more goodies in the paper
doi.org/10.1101/2024...

We find that much of the variation can be explained and predicted by a burial-dependent substitution matrix
Lots more goodies in the paper
doi.org/10.1101/2024...
www1.bio.ku.dk/nyheder/2024...

www1.bio.ku.dk/nyheder/2024...

