
https://linktr.ee/shiori_iida
Using the RL algorithm (from pnas.org/doi/10.1073/...),
@masaashimazoe.bsky.social analyzed our nucleosome tracking data and classified them by their mobility.
Hotter color = faster motion (Slow🔵→ 🟢 → 🟡 → 🟠 Fast)
www.science.org/doi/10.1126/...
www.science.org/doi/10.1126/...

www.umass.edu/biology/abou...
www.umass.edu/biology/abou...
I’ll keep doing exciting science with joy and passion 🔬💫
Huge thanks to @kazu-maeshima.bsky.social lab for all the support during my PhD journey⛰️
I’ll start by tasting every single sausage here🇩🇪
I’ll keep doing exciting science with joy and passion 🔬💫
Huge thanks to @kazu-maeshima.bsky.social lab for all the support during my PhD journey⛰️
I’ll start by tasting every single sausage here🇩🇪
I’ll keep doing exciting science with joy and passion 🔬💫
Huge thanks to @kazu-maeshima.bsky.social lab for all the support during my PhD journey⛰️
I’ll start by tasting every single sausage here🇩🇪

www.biorxiv.org/cgi/content/...
www.biorxiv.org/cgi/content/...
With @shiori-iida.bsky.social , We revealed the detailed structure of euchromatin using super-resolution imaging.
Cohesin regulates the function of euchromatin via chromatin dynamics!
(1/n)
www.biorxiv.org/cgi/content/...
With @shiori-iida.bsky.social , We revealed the detailed structure of euchromatin using super-resolution imaging.
Cohesin regulates the function of euchromatin via chromatin dynamics!
(1/n)
Using the RL algorithm (from pnas.org/doi/10.1073/...),
@masaashimazoe.bsky.social analyzed our nucleosome tracking data and classified them by their mobility.
Hotter color = faster motion (Slow🔵→ 🟢 → 🟡 → 🟠 Fast)
Using the RL algorithm (from pnas.org/doi/10.1073/...),
@masaashimazoe.bsky.social analyzed our nucleosome tracking data and classified them by their mobility.
Hotter color = faster motion (Slow🔵→ 🟢 → 🟡 → 🟠 Fast)
We combined single-nucleosome imaging and 3D-SIM to reveal:
🔹 Euchromatin forms condensed domains, not open fibers
🔹Cohesin loss increases nucleosome mobility without decompaction
🔹Cohesin prevents neighboring domain mixing
Full story & movies👇
www.biorxiv.org/cgi/content/...
We combined single-nucleosome imaging and 3D-SIM to reveal:
🔹 Euchromatin forms condensed domains, not open fibers
🔹Cohesin loss increases nucleosome mobility without decompaction
🔹Cohesin prevents neighboring domain mixing
Full story & movies👇
💚 Thanks to the team @biswashere.bsky.social, Omar Muñoz, ✨Q✨ C. Hoege, B. Lorton, R. Nikolay @matthewkraushar.bsky.social @dshechter.bsky.social @gucklab.bsky.social @vasilyzaburdaev.bsky.social 💚

💚 Thanks to the team @biswashere.bsky.social, Omar Muñoz, ✨Q✨ C. Hoege, B. Lorton, R. Nikolay @matthewkraushar.bsky.social @dshechter.bsky.social @gucklab.bsky.social @vasilyzaburdaev.bsky.social 💚
Link: biorxiv.org/content/10.110…
Using single-molecul#imagingng, we showed that liquid-like BRD4-NUT condensates locally constrai#chromatinin vi#bromodomain-dependentnt crosslinking.
I thank my supervisor, Prof. @kazu-maeshima.bsky.social, for his support!

Link: biorxiv.org/content/10.110…
Using single-molecul#imagingng, we showed that liquid-like BRD4-NUT condensates locally constrai#chromatinin vi#bromodomain-dependentnt crosslinking.
I thank my supervisor, Prof. @kazu-maeshima.bsky.social, for his support!
BRD4-NUT forms liquid-like condensates that locally constrain nucleosomes via BRD4-mediated crosslinking— physical control of #chromatin by #LLPS transcription condensates. @semeigazin.bsky.social @katsuminami.bsky.social @masaashimazoe.bsky.social
BRD4-NUT forms liquid-like condensates that locally constrain nucleosomes via BRD4-mediated crosslinking— physical control of #chromatin by #LLPS transcription condensates. @semeigazin.bsky.social @katsuminami.bsky.social @masaashimazoe.bsky.social
www.nig.ac.jp/nig/career-d...
🧬We’re seeking motivated postdocs to join our lab. 🔬We study chromatin behavior in living cells using advanced live-cell imaging and quantitative analysis:
www.science.org/doi/10.1126/...
doi.org/10.2183/pjab...
Interested? Please apply!

www.nig.ac.jp/nig/career-d...
🧬We’re seeking motivated postdocs to join our lab. 🔬We study chromatin behavior in living cells using advanced live-cell imaging and quantitative analysis:
www.science.org/doi/10.1126/...
doi.org/10.2183/pjab...
Interested? Please apply!
Our new manuscript is now available as a preprint!⛅️
biorxiv.org/content/10.1...
1/ Here, we describe a novel BMAL1-YAP complex in epidermal cells, which is hijacked during ageing to control the expression of up-regulated inflammation genes through enhancer binding.🧵

🔬 Original paper: www.science.org/doi/full/10....

🔬 Original paper: www.science.org/doi/full/10....
doi.org/10.2183/pjab...
The textbook view of #chromatin as 30-nm fibers is fading.
A new paradigm: chromatin forms #liquid-like domains—locally dynamic, yet globally stable at the chromosome level (viscoelastic)—supporting genome function. ✨
doi.org/10.2183/pjab...
The textbook view of #chromatin as 30-nm fibers is fading.
A new paradigm: chromatin forms #liquid-like domains—locally dynamic, yet globally stable at the chromosome level (viscoelastic)—supporting genome function. ✨
www.science.org/doi/10.1126/...
🧬Our Repli-Histo labeling marks nucleosomes in euchromatin and heterochromatin in live human cells.
🔍 @katsuminami.bsky.social et al. have developed a chromatin behavior atlas within the nucleus. 1/2
www.science.org/doi/10.1126/...
🧬Our Repli-Histo labeling marks nucleosomes in euchromatin and heterochromatin in live human cells.
🔍 @katsuminami.bsky.social et al. have developed a chromatin behavior atlas within the nucleus. 1/2
I can't thank @kazu-maeshima.bsky.social, my labmates, and all the NIG members enough for their support.
This recognition gives me even more motivation for the journey ahead!


I can't thank @kazu-maeshima.bsky.social, my labmates, and all the NIG members enough for their support.
This recognition gives me even more motivation for the journey ahead!
Huge thanks to everyone supported me, especially my mentor @kazu-maeshima.bsky.social, labmates, friends, and family.
I’m excited to keep chasing big questions and growing as a scientist!

Huge thanks to everyone supported me, especially my mentor @kazu-maeshima.bsky.social, labmates, friends, and family.
I’m excited to keep chasing big questions and growing as a scientist!
@masaashimazoe.bsky.social et al. reveal that linker histone H1 acts as a liquid-like glue to organize chromatin in living cells. 🎉 Fantastic collab with @rcollepardo.bsky.social @janhuemar.bsky.social and others—huge thanks! 🙌 1/
@masaashimazoe.bsky.social et al. reveal that linker histone H1 acts as a liquid-like glue to organize chromatin in living cells. 🎉 Fantastic collab with @rcollepardo.bsky.social @janhuemar.bsky.social and others—huge thanks! 🙌 1/
We critically discuss the domain formation mechanism from a physical perspective, including #phase-separation and #condensation.
📥 Free-download link:
authors.elsevier.com/a/1keGn,LqAr...
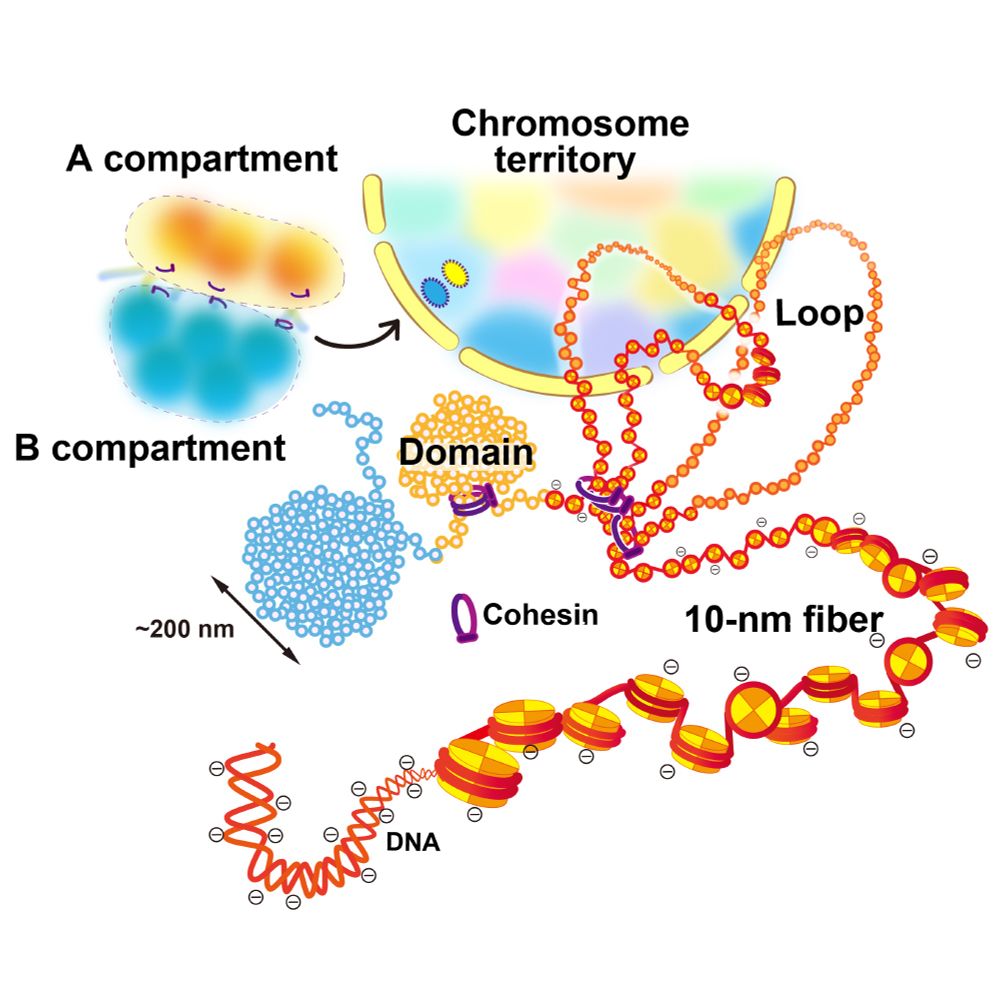
We critically discuss the domain formation mechanism from a physical perspective, including #phase-separation and #condensation.
📥 Free-download link:
authors.elsevier.com/a/1keGn,LqAr...
#chromatin #genomics
www.science.org/doi/10.1126/...

#chromatin #genomics
www.science.org/doi/10.1126/...


