

These papers describe a remarkable role for *recessive* variants in *RNU2-2* causing developmental and epileptic encephalopathy
🧵 by the amazing @christeldepienne.bsky.social 👇
1/3
📄 www.medrxiv.org/content/10.1...

These papers describe a remarkable role for *recessive* variants in *RNU2-2* causing developmental and epileptic encephalopathy
🧵 by the amazing @christeldepienne.bsky.social 👇
1/3
www.medrxiv.org/content/10.1...

www.medrxiv.org/content/10.1...
If you're interested in a PhD, check out our department @cam.ac.uk www.phpc.cam.ac.uk/education-an...
We've a super interdisciplinary faculty spanning primary care, data science, epidemiology etc
If you're interested in a PhD, check out our department @cam.ac.uk www.phpc.cam.ac.uk/education-an...
We've a super interdisciplinary faculty spanning primary care, data science, epidemiology etc
We describe the clinical phenotype of a recessive NDD associated with biallelic variants in RNU4-2 🧬
See 🧵👇
tinyurl.com/3j9r56s8
@rociorius.bsky.social @yuyangchen.bsky.social @gregfindlay.bsky.social @dgmacarthur.bsky.social @cassimons.bsky.social @nickywhiffin.bsky.social

We describe the clinical phenotype of a recessive NDD associated with biallelic variants in RNU4-2 🧬
See 🧵👇
Maternal and paternal alleles can have distinct — even opposite — effects on human traits, revealing a hidden layer of genetic architecture that standard GWAS miss.
🔗 www.nature.com/articles/s41...
Highlights below!
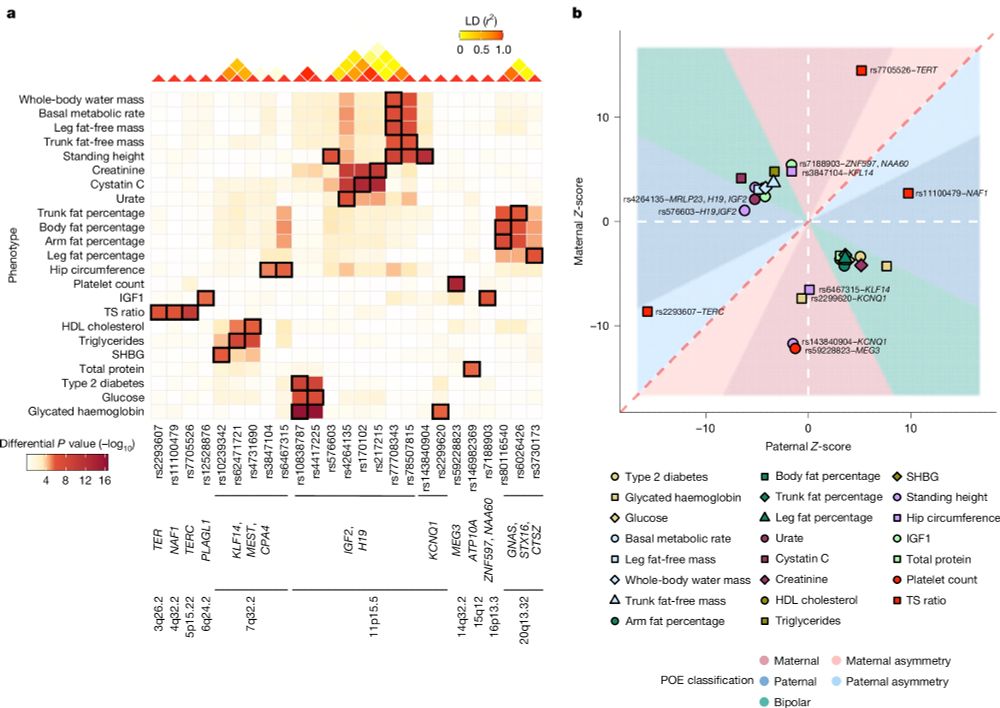
Maternal and paternal alleles can have distinct — even opposite — effects on human traits, revealing a hidden layer of genetic architecture that standard GWAS miss.
🔗 www.nature.com/articles/s41...
Highlights below!
Check out our Green Algorithms Initiative for more tools and information www.green-algorithms.org
And the GREENER Principles which lay the foundation for a sustainability roadmap www.nature.com/articles/s43...
Check out our Green Algorithms Initiative for more tools and information www.green-algorithms.org
And the GREENER Principles which lay the foundation for a sustainability roadmap www.nature.com/articles/s43...
Cael y diweddaraf am ddatblygiadau a straeon ysbrydoledig sy’n siapio dyfodol #arloesi ym maes #iechyd a #GofalCymdeithasol yng Nghymru a’r tu hwnt: www.linkedin.com/feed/update/...

Cael y diweddaraf am ddatblygiadau a straeon ysbrydoledig sy’n siapio dyfodol #arloesi ym maes #iechyd a #GofalCymdeithasol yng Nghymru a’r tu hwnt: www.linkedin.com/feed/update/...
Tanysgrifiwch i Hwb Heddiw: lshubwales.com/cy/our-newsl...

Tanysgrifiwch i Hwb Heddiw: lshubwales.com/cy/our-newsl...
Subscribe to Hub Highlights today: lshubwales.com/our-newsletter

Subscribe to Hub Highlights today: lshubwales.com/our-newsletter
Stay in the loop with developments, projects, and inspiring stories shaping the future of #health and #SocialCare #innovation across Wales and beyond: www.linkedin.com/feed/update/...

Stay in the loop with developments, projects, and inspiring stories shaping the future of #health and #SocialCare #innovation across Wales and beyond: www.linkedin.com/feed/update/...
Workshop materials: biocpy.github.io/BiocWorkshop...
Book: biocpy.github.io/tutorial/
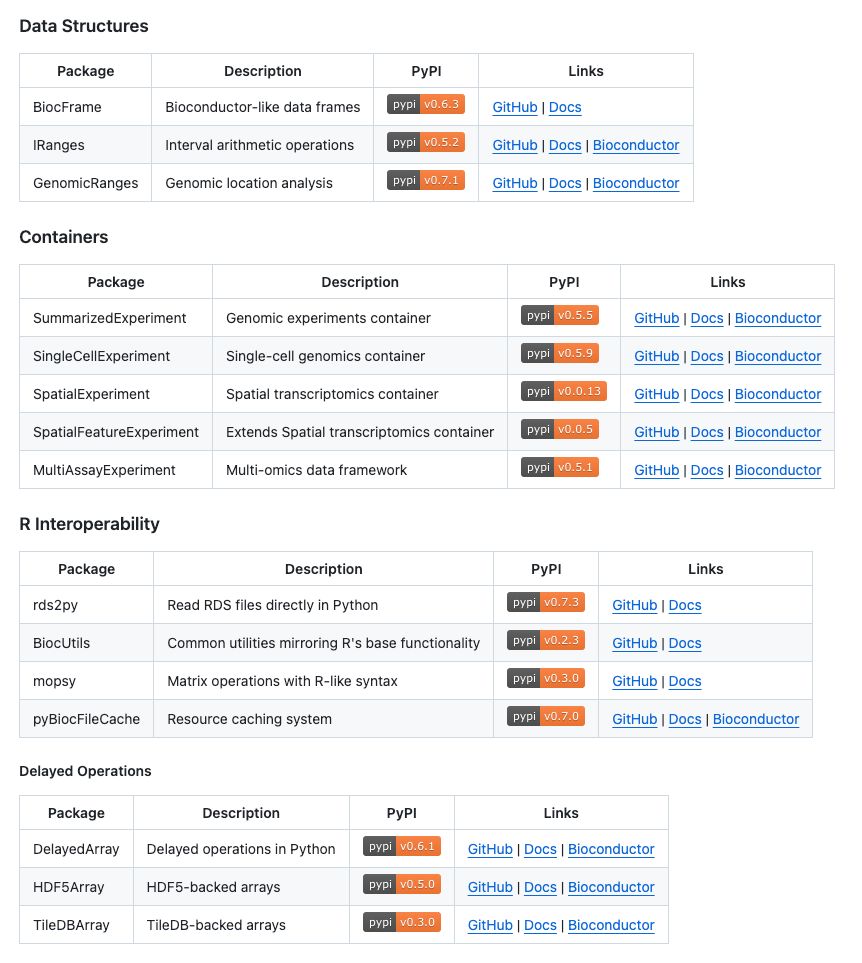
Workshop materials: biocpy.github.io/BiocWorkshop...
Book: biocpy.github.io/tutorial/
GW is a fast genomics browser (up to 100x faster!)
github.com/kcleal/gw
Also, just released a Python interface for GW
github.com/kcleal/gwplot
📝 nature.com/articles/s4159…#Genomicsc#Bioinformaticscs
GW is a fast genomics browser (up to 100x faster!)
github.com/kcleal/gw
Also, just released a Python interface for GW
github.com/kcleal/gwplot
📝 nature.com/articles/s4159…#Genomicsc#Bioinformaticscs

See the advert here:
krb-sjobs.brassring.com/TGnewUI/Search…
And contact WAVU co-director David Gillespie for informal enquiries.
#NewPI #Lecturer #GroupLeader #Job #Opportunity
See the advert here:
krb-sjobs.brassring.com/TGnewUI/Search…
And contact WAVU co-director David Gillespie for informal enquiries.
#NewPI #Lecturer #GroupLeader #Job #Opportunity
www.medrxiv.org/content/10.1...
A super fun collaboration with incredible duo @gregfindlay.bsky.social @joachimdejonghe.bsky.social from @crick.ac.uk
🧬🖥️🩺
🧵1/12

www.medrxiv.org/content/10.1...
A super fun collaboration with incredible duo @gregfindlay.bsky.social @joachimdejonghe.bsky.social from @crick.ac.uk
🧬🖥️🩺
🧵1/12

github.com/lcpilling/uk...
github.com/lcpilling/uk...
Check it out: github.com/chhylp123/hi...


