


GW is a fast genomics browser (up to 100x faster!)
github.com/kcleal/gw
Also, just released a Python interface for GW
github.com/kcleal/gwplot
📝 nature.com/articles/s4159…#Genomicsc#Bioinformaticscs
GW is a fast genomics browser (up to 100x faster!)
github.com/kcleal/gw
Also, just released a Python interface for GW
github.com/kcleal/gwplot
📝 nature.com/articles/s4159…#Genomicsc#Bioinformaticscs
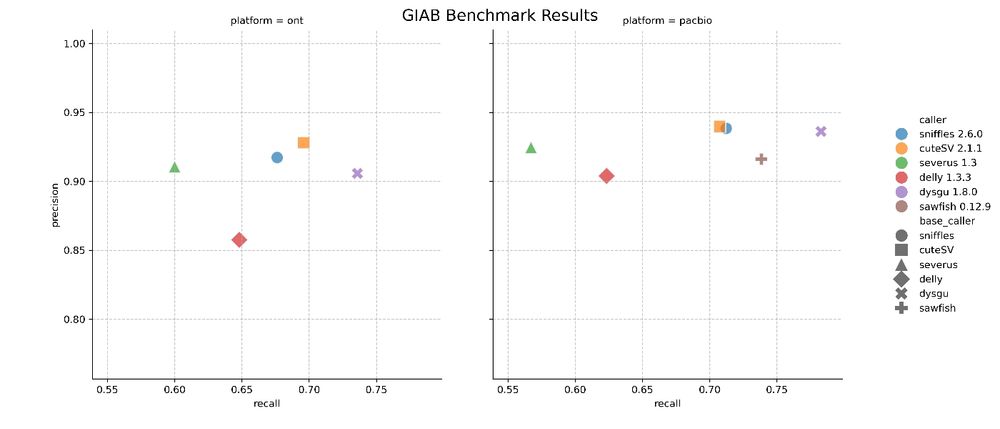
go.bsky.app/TRWCnZs
go.bsky.app/TRWCnZs



