| 🧬 Researching the evolution of antimicrobial resistance
| 🦠 Microbiology, Genomics, Bioinformatics
We used comparative genomics to determine the relatedness of E. coli and K. pneumoniae isolates from faeces and blood of the same neonates with bloodstream infections.
doi.org/10.1038/s420...

We used comparative genomics to determine the relatedness of E. coli and K. pneumoniae isolates from faeces and blood of the same neonates with bloodstream infections.
doi.org/10.1038/s420...
www.nature.com/articles/s42...

www.nature.com/articles/s42...

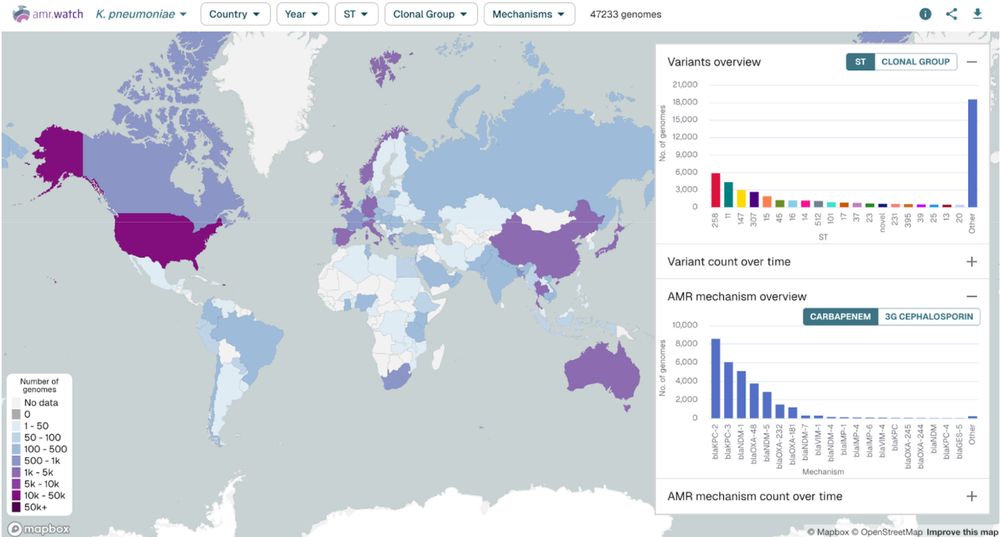
@lstmnews.bsky.social @gcagatgcaatg.bsky.social @russstothard.bsky.social
www.biorxiv.org/content/10.1...

@lstmnews.bsky.social @gcagatgcaatg.bsky.social @russstothard.bsky.social
www.biorxiv.org/content/10.1...
go.bsky.app/EUY2FUJ
Follow these amazing people along with those in Vol. 1
go.bsky.app/KkZfvrh
And don't forget to pin the Antimicrobial Resistance feed to get all the AMR posts in one place
bsky.app/profile/did:...
go.bsky.app/EUY2FUJ
Follow these amazing people along with those in Vol. 1
go.bsky.app/KkZfvrh
And don't forget to pin the Antimicrobial Resistance feed to get all the AMR posts in one place
bsky.app/profile/did:...
I'm excited to announce Autocycler, my new tool for consensus assembly of long-read bacterial genomes!
It's the successor to Trycycler, designed to be faster and less reliant on user intervention.
Check it out: github.com/rrwick/Autoc...
(1/5)
I'm excited to announce Autocycler, my new tool for consensus assembly of long-read bacterial genomes!
It's the successor to Trycycler, designed to be faster and less reliant on user intervention.
Check it out: github.com/rrwick/Autoc...
(1/5)
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
@naturecomms.bsky.social
.
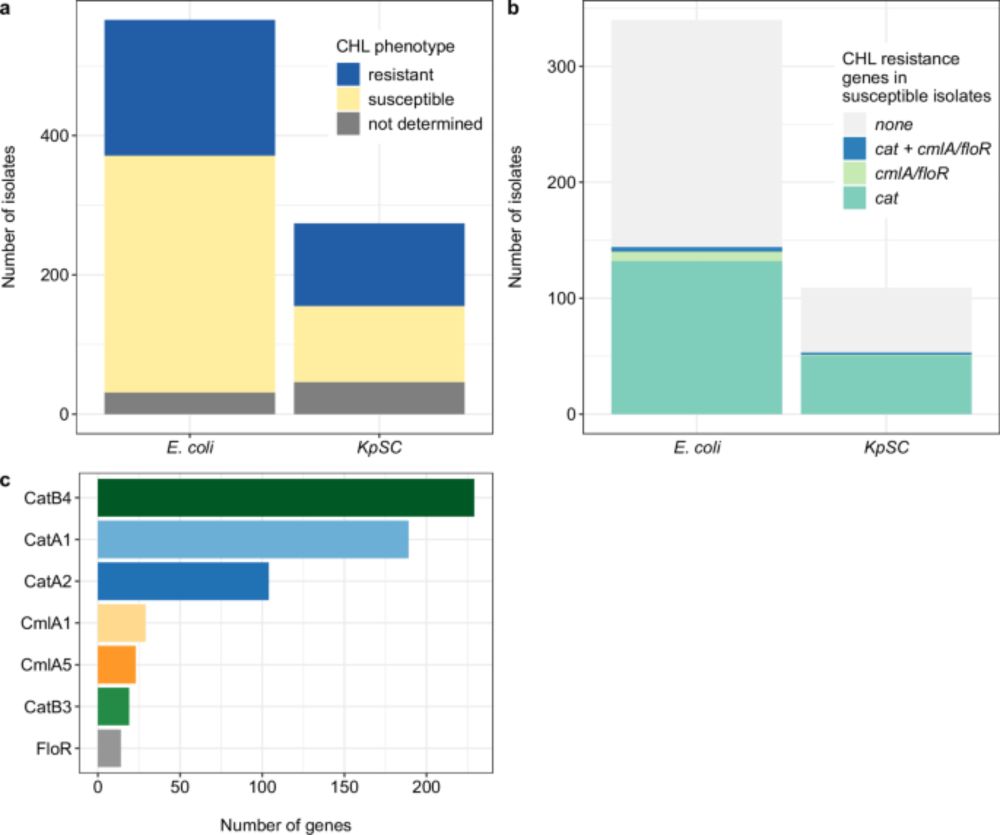
Here we use various techniques to determine the molecular mechanisms for an observed rise in chloramphenicol susceptibility at the same time as an increase in ESBL resistance.
@naturecomms.bsky.social
.
Here we use various techniques to determine the molecular mechanisms for an observed rise in chloramphenicol susceptibility at the same time as an increase in ESBL resistance.

