| 🧬 Researching the evolution of antimicrobial resistance
| 🦠 Microbiology, Genomics, Bioinformatics
This was a great team effort led by Fabrice Graf from LSTM and it's great to see it finally published!
This was a great team effort led by Fabrice Graf from LSTM and it's great to see it finally published!
(i) that antibiotic susceptibility can re-emerge after an antibiotic stops being used
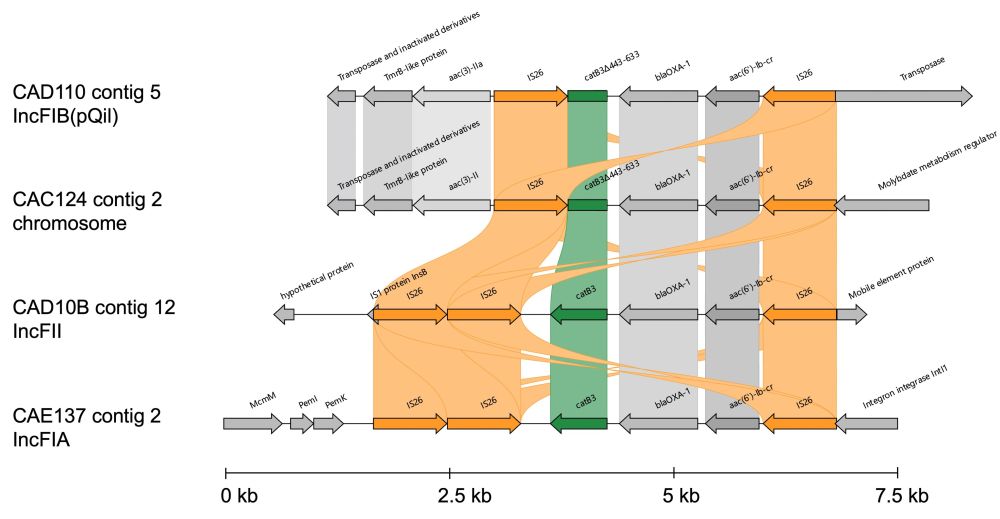
(ii) that genetic context should be included in AMR databases
(iii) that chloramphenicol could be reintroduced in certain contexts (e.g critically ill with ESBL-E)
6/7
(i) that antibiotic susceptibility can re-emerge after an antibiotic stops being used
(ii) that genetic context should be included in AMR databases
(iii) that chloramphenicol could be reintroduced in certain contexts (e.g critically ill with ESBL-E)
6/7
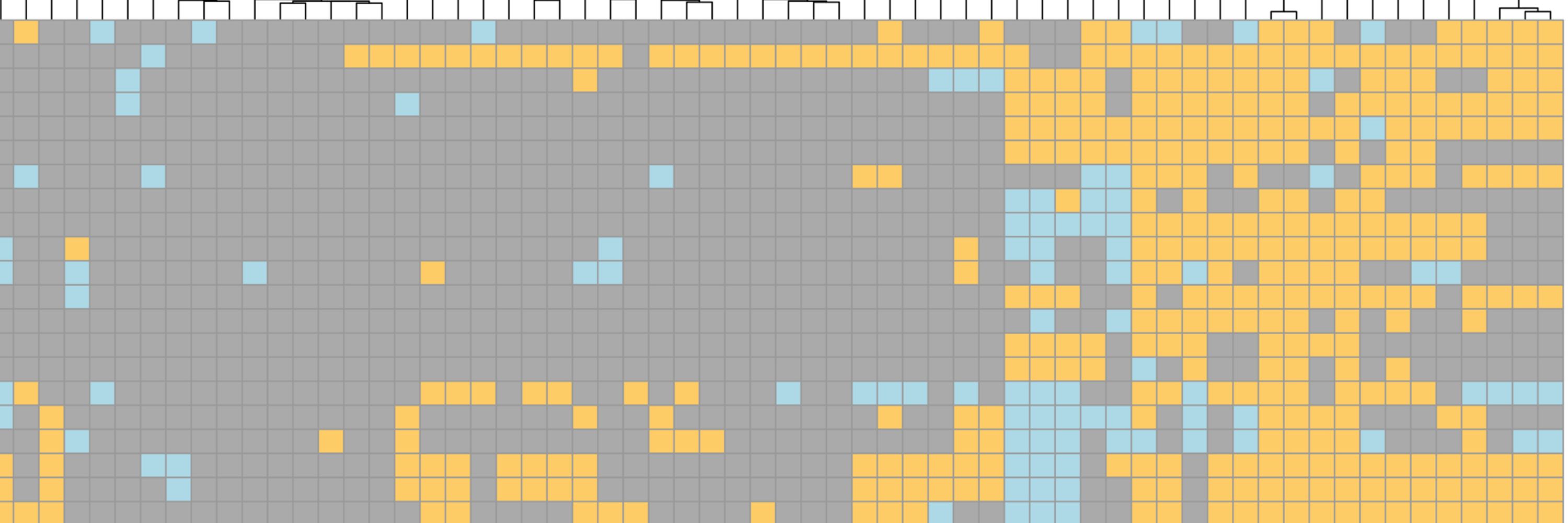
5/7
5/7
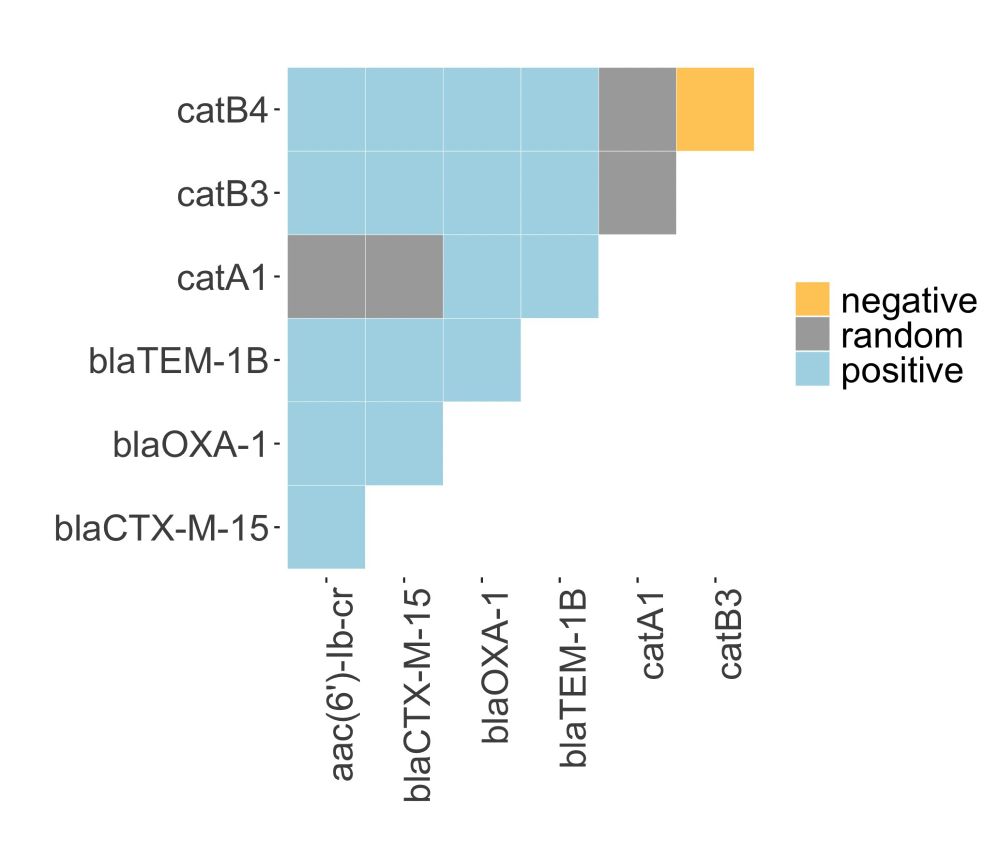
4/7
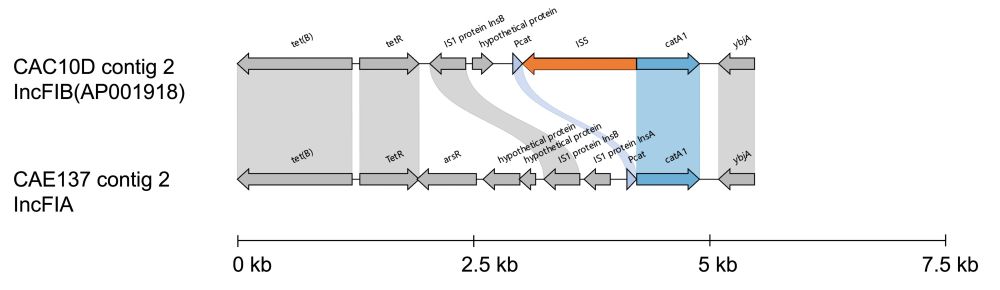
4/7
3/7
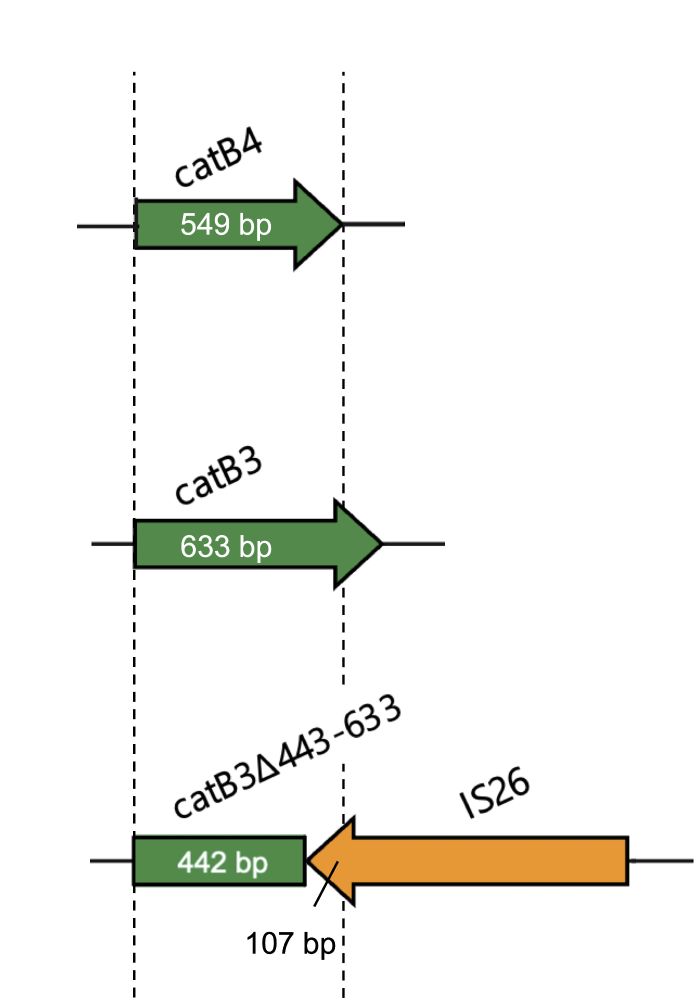
3/7
2/7
2/7

