Interests: Protein Quality Control, Genetics, Molecular Chaperones, Degrons.
Martin Grønbæk-Thygesen (from @rhp-lab.bsky.social) et al show that introducing K here causes ubiquitylation and degradation
doi.org/10.1101/2025...

Martin Grønbæk-Thygesen (from @rhp-lab.bsky.social) et al show that introducing K here causes ubiquitylation and degradation
doi.org/10.1101/2025...
doi.org/10.1038/s414...

doi.org/10.1038/s414...
I thought science hinged on prestige. Moving abroad made me reassess my priorities
www.science.org/content/arti...

I thought science hinged on prestige. Moving abroad made me reassess my priorities
www.science.org/content/arti...
Register to attend:
tinyurl.com/PMCseminar2
Recordings of previous seminars: www.youtube.com/@PMCModularity

Register to attend:
tinyurl.com/PMCseminar2
Recordings of previous seminars: www.youtube.com/@PMCModularity
efzu.fa.em2.oraclecloud.com/hcmUI/Candid...

efzu.fa.em2.oraclecloud.com/hcmUI/Candid...
www.ft.com/content/25dd...
www.ft.com/content/25dd...
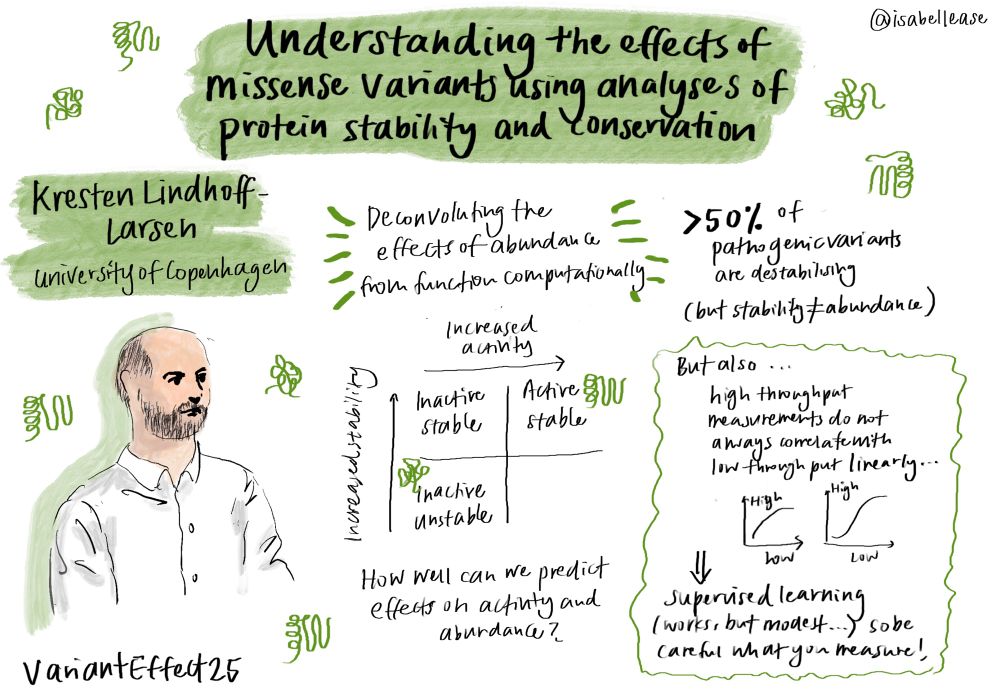


☘️ mechanisms of #SAM homeostasis (Ben Tu et al)
☘️ glucose sensing by #glucokinase and how disease variants impact its function @sarahgersing.bsky.social @lindorfflarsen.bsky.social @rhp-lab.bsky.social
☘️ mechanisms of #SAM homeostasis (Ben Tu et al)
☘️ glucose sensing by #glucokinase and how disease variants impact its function @sarahgersing.bsky.social @lindorfflarsen.bsky.social @rhp-lab.bsky.social
#GlucoseHomeostasis #GlucoseSensor #GCKMODY #CooperativeKinetics
authors.elsevier.com/a/1kN5M3S6Gf...
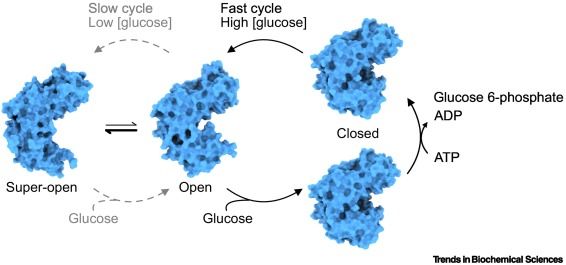
#GlucoseHomeostasis #GlucoseSensor #GCKMODY #CooperativeKinetics
authors.elsevier.com/a/1kN5M3S6Gf...
Link to paper: www.cell.com/structure/fu...
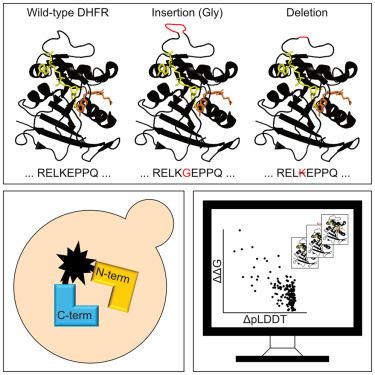
Link to paper: www.cell.com/structure/fu...

