


Then check out @sarahgersing.bsky.social's review on glucokinase: doi.org/10.1016/j.ti...
☘️ mechanisms of #SAM homeostasis (Ben Tu et al)
☘️ glucose sensing by #glucokinase and how disease variants impact its function @sarahgersing.bsky.social @lindorfflarsen.bsky.social @rhp-lab.bsky.social
Then check out @sarahgersing.bsky.social's review on glucokinase: doi.org/10.1016/j.ti...
#GlucoseHomeostasis #GlucoseSensor #GCKMODY #CooperativeKinetics
authors.elsevier.com/a/1kN5M3S6Gf...
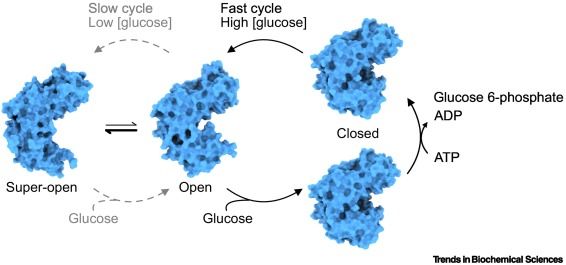
#GlucoseHomeostasis #GlucoseSensor #GCKMODY #CooperativeKinetics
authors.elsevier.com/a/1kN5M3S6Gf...
Link to paper: www.cell.com/structure/fu...
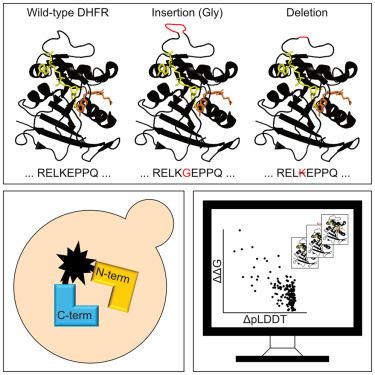
Link to paper: www.cell.com/structure/fu...

