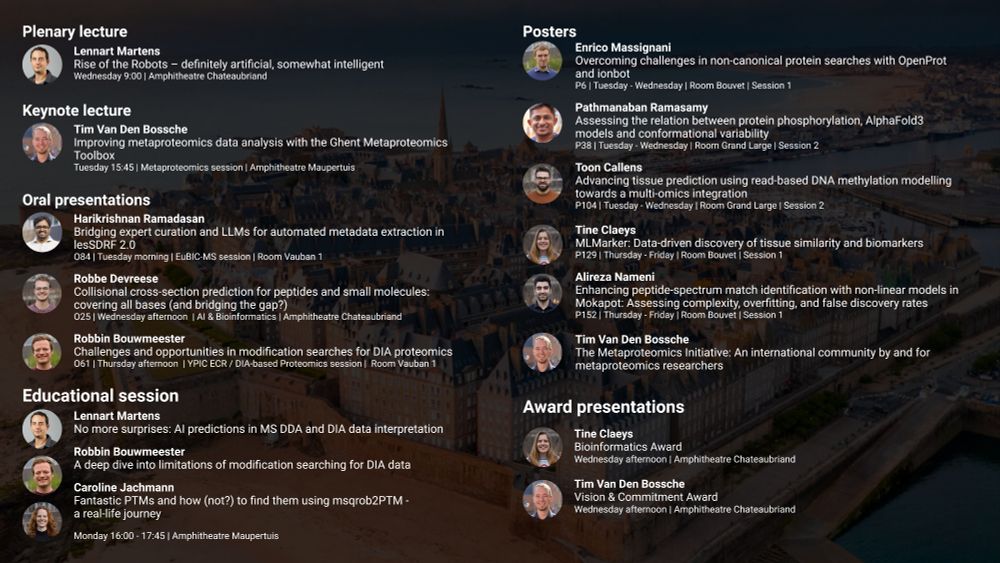
Proteomics, Mass Spectrometry, Bioinformatics, Machine learning
#Proteomics #MassSpec
🧵 ½

#Proteomics #MassSpec
🧵 ½
Join us in Harrachov, Czechia for a week of keynotes, workshops, and networking on computational MS.
Info and registration: eubic-ms.org/events/2026-...
#EuBIC2026 #MassSpectrometry #MassSpec #Bioinformatics #Proteomics #Metabolomics

👉 Learn more about our open proteomics software benchmarking platform at proteobench.readthedocs.io.
#Proteomics #Bioinformatics #Benchmarking

👉 Learn more about our open proteomics software benchmarking platform at proteobench.readthedocs.io.
#Proteomics #Bioinformatics #Benchmarking
It's like the philosopher's stone, turning your experimental data into gold and giving it the elixir of life.
Congrats with the EuPA Bioinformatics Award, Tine!
#EuPA2025

It's like the philosopher's stone, turning your experimental data into gold and giving it the elixir of life.
Congrats with the EuPA Bioinformatics Award, Tine!
#EuPA2025
www.matrixscience.com/blog/using-m...
www.matrixscience.com/blog/using-m...
www.matrixscience.com/blog/using-m...
www.matrixscience.com/blog/using-m...
Check it out here:
www.biorxiv.org/content/10.1...

Check it out here:
www.biorxiv.org/content/10.1...
Our latest research, TIMS²Rescore, is now published in Journal of Proteome Research! 🎉
Read it here: pubs.acs.org/doi/full/10....
A huge thanks to all our collaborators for making this happen!

Our latest research, TIMS²Rescore, is now published in Journal of Proteome Research! 🎉
Read it here: pubs.acs.org/doi/full/10....
A huge thanks to all our collaborators for making this happen!
#EuBIC2025




#EuBIC2025
How do we unlock proteomics data reuse, tackle metadata challenges, and harness public (clinical) data for AI?
Find out at HUPO-PSI Spring Meeting 2025!
📅 March 31 – April 3, 2025
📍 Tübingen, Germany
(1/4)
How do we unlock proteomics data reuse, tackle metadata challenges, and harness public (clinical) data for AI?
Find out at HUPO-PSI Spring Meeting 2025!
📅 March 31 – April 3, 2025
📍 Tübingen, Germany
(1/4)


A big thanks to Louise Buur and Viktoria Dorfer for the great collaboration!
Open access full text: biblio.ugent.be/publication/...
Project: github.com/compomics/ms...
A big thanks to Louise Buur and Viktoria Dorfer for the great collaboration!
Open access full text: biblio.ugent.be/publication/...
Project: github.com/compomics/ms...
1. Bruker has the greatest momentum with a 4-fold increase since 2017
- timsTOFs are largely responsible for this increase
2. Sciex is gradually losing market share

1. Bruker has the greatest momentum with a 4-fold increase since 2017
- timsTOFs are largely responsible for this increase
2. Sciex is gradually losing market share
proteomecentral.proteomexchange.org/usi/?usi=mzs...
proteomecentral.proteomexchange.org/usi/?usi=mzs...
EuBIC-MS community members did, and started #ProteoBench: An open platform for comparing proteomics data analysis workflows.
Learn more at the webinar (27 Feb, 18h CET) Register here: bit.ly/3SfS4Lf
EuBIC-MS community members did, and started #ProteoBench: An open platform for comparing proteomics data analysis workflows.
Learn more at the webinar (27 Feb, 18h CET) Register here: bit.ly/3SfS4Lf
user-friendly platform to boost peptide
identifications, as showcased with MS Amanda 3.0 chemrxiv.org/engage/...
---
#proteomics #prot-preprint

---
#proteomics #prot-paper




Registration info on eubic-ms.org/events/2024-...

Registration info on eubic-ms.org/events/2024-...