Views my own.
Mastodon @Psy_Fer_@genomic.social, https://genomic.social
I had to write a function that found the factors of a number. I was about to write a prime shortcut function too, but remembered the kmer lengths wouldn't go above 62, so i could just store those in a vector.
I had to write a function that found the factors of a number. I was about to write a prime shortcut function too, but remembered the kmer lengths wouldn't go above 62, so i could just store those in a vector.
Highly relatable for anyone that has ever written a line of code used by other people
Lovely little post from @jacobtomlinson.dev

Highly relatable for anyone that has ever written a line of code used by other people
Lovely little post from @jacobtomlinson.dev

It's the first tool that builds on Sassy, the approximate-DNA-searching tool that @rickbitloo.bsky.social and myself developed earlier this year, specifically with this application in mind.
www.biorxiv.org/content/10.1...
Want to get started? github.com/rickbeeloo/b...
It's the first tool that builds on Sassy, the approximate-DNA-searching tool that @rickbitloo.bsky.social and myself developed earlier this year, specifically with this application in mind.
I did this because the r/bioinformatics channel doesn't like talking about tools, especially if you wrote it yourself.
If you build bioinformatic tools or want to learn more about it, join the subreddit and start posting :)

I did this because the r/bioinformatics channel doesn't like talking about tools, especially if you wrote it yourself.
If you build bioinformatic tools or want to learn more about it, join the subreddit and start posting :)
How about the matching coordinates of an assembly mapped to a reference but repeats and SVs are confusing liftover?
Well I did while trying to benchmark STR stuff. Nice side quest.
github.com/Psy-Fer/bedp...

How about the matching coordinates of an assembly mapped to a reference but repeats and SVs are confusing liftover?
Well I did while trying to benchmark STR stuff. Nice side quest.
github.com/Psy-Fer/bedp...
@GigaScience
academic.oup.com/gigascience/....
Some plots required a log scale - RAM usage and random access time.

@GigaScience
academic.oup.com/gigascience/....
Some plots required a log scale - RAM usage and random access time.
docs.google.com/document/d/1...

docs.google.com/document/d/1...
AAMRI/KPMG found that every $1 invested in Australian medical research returns $3.90 to the economy
AAMRI/KPMG found that every $1 invested in Australian medical research returns $3.90 to the economy
#NHMRCdraftStratergy #Ihavenotes #ValueFundamentalScience
#NHMRCdraftStratergy #Ihavenotes #ValueFundamentalScience
So I'm going to...🧵🤔🧵😱🧵😵💫🧵🙏🧵🧵🧵
This is a repost from X because you all need to know!
#NHMRCdraftStratergy #Ihavenotes #ValueFundamentalScience
@nhmrc.bsky.social
So I'm going to...🧵🤔🧵😱🧵😵💫🧵🙏🧵🧵🧵
This is a repost from X because you all need to know!
#NHMRCdraftStratergy #Ihavenotes #ValueFundamentalScience
@nhmrc.bsky.social
tinyurl.com/4d337djb

tinyurl.com/4d337djb

Also for being awarded the #EurekaPrize for outstanding early career researcher, showcase his pioneering and impacftful research in #bioinformatics software


Also for being awarded the #EurekaPrize for outstanding early career researcher, showcase his pioneering and impacftful research in #bioinformatics software

It was pretty easy to refactor, with compiler/rust analyser. Love this language
It was pretty easy to refactor, with compiler/rust analyser. Love this language
@nanoporetech.com
basecalling
@petermaccc.bsky.social
Threadripper
4x5090, liquid cooled
128gb DDR5 ram
~40Tb of nvme and SSD storage
TWO power supplies, a 1200W and a 2500W
All in a Corsair 9000D case
~5^7 samples/second with dorado.

@nanoporetech.com
basecalling
@petermaccc.bsky.social
Threadripper
4x5090, liquid cooled
128gb DDR5 ram
~40Tb of nvme and SSD storage
TWO power supplies, a 1200W and a 2500W
All in a Corsair 9000D case
~5^7 samples/second with dorado.
www.eventbrite.com.au/e/garvan-lon...
www.eventbrite.com.au/e/garvan-lon...
1.79e+07 samples/s
That's ~123gbp/day
Wow!
@nanoporetech.com have done a great job with the speedups.

1.79e+07 samples/s
That's ~123gbp/day
Wow!
@nanoporetech.com have done a great job with the speedups.
biorxiv.org/content/10.1...
-similar accuracy to modkit & pb-CpG-tools.
-standard open-source licenses (NOT vendor-specific)
-Simple but faster, on a laptop ~4X for DNA and ~55X for RNA.
Code: github.com/warp9seq/min...
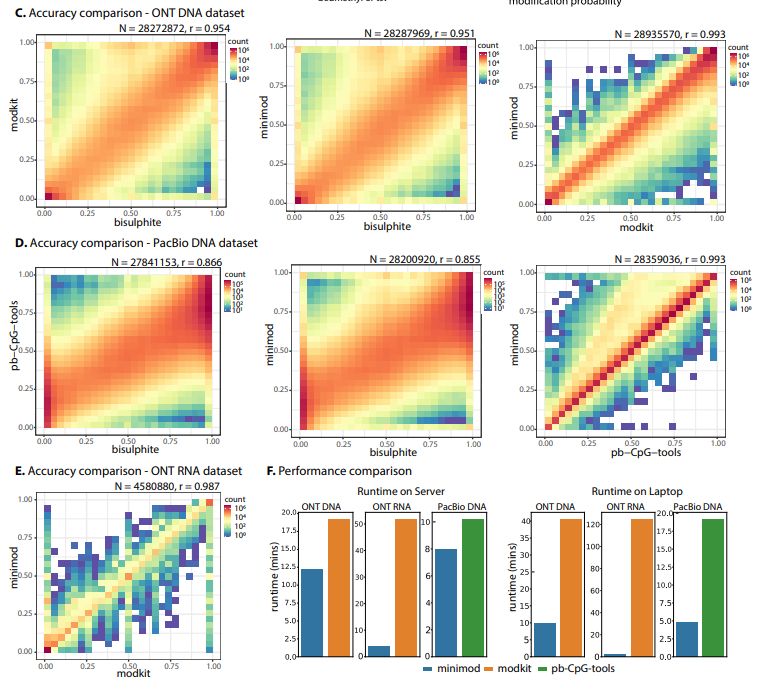
biorxiv.org/content/10.1...
-similar accuracy to modkit & pb-CpG-tools.
-standard open-source licenses (NOT vendor-specific)
-Simple but faster, on a laptop ~4X for DNA and ~55X for RNA.
Code: github.com/warp9seq/min...
@bosc.bsky.social around 2:30pm ish where
@sunethsa.bsky.social will present real-time @nanoporetech.com frequency calculation using realfreq & standalone frequency calculation using minimod.
academic.oup.com/bioinformati...
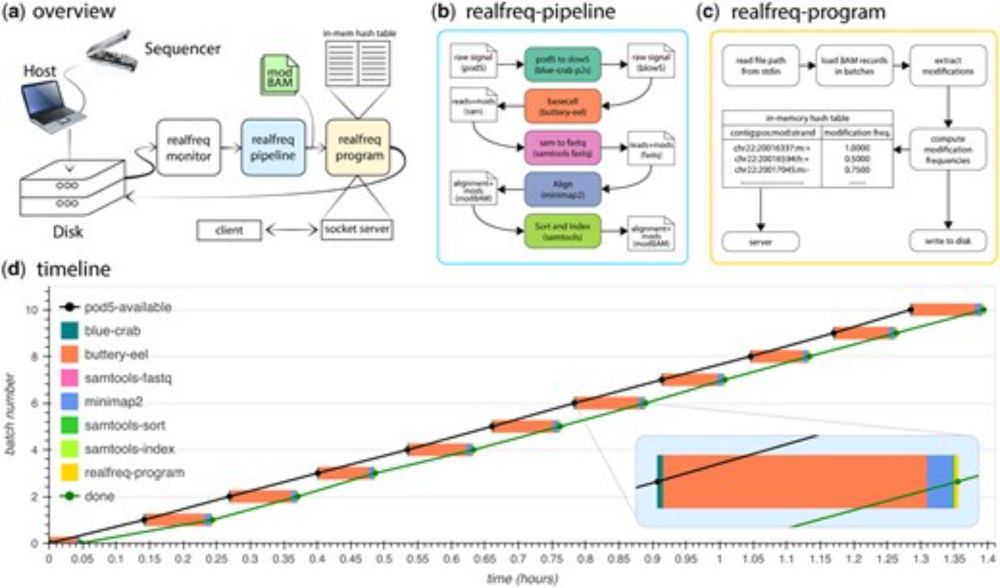
@bosc.bsky.social around 2:30pm ish where
@sunethsa.bsky.social will present real-time @nanoporetech.com frequency calculation using realfreq & standalone frequency calculation using minimod.
academic.oup.com/bioinformati...
@bosc.bsky.social track around 2:30pm ish after
@sunethsa.bsky.social's talk, Bonson Wong will present on
nanopore basecalling on AMD GPUs using slorado
github.com/BonsonW/slor...
@bosc.bsky.social track around 2:30pm ish after
@sunethsa.bsky.social's talk, Bonson Wong will present on
nanopore basecalling on AMD GPUs using slorado
github.com/BonsonW/slor...
github.com/warp9seq/min...
written by
@sunethsa.bsky.social
in C, based on mod tag parsing we did for realfreq doi.org/10.1093/bioi...
github.com/warp9seq/min...
written by
@sunethsa.bsky.social
in C, based on mod tag parsing we did for realfreq doi.org/10.1093/bioi...

