Designing embedded systems for bioinformatics applications.
GitHub: github.com/hasindu2008/...
Paper: nature.com/articles/s41...
Datasets: hasindu2008.github.io/cornetto/doc...
... and a banner made by Ira Deveson referring to ‘no boring bits’.

GitHub: github.com/hasindu2008/...
Paper: nature.com/articles/s41...
Datasets: hasindu2008.github.io/cornetto/doc...
... and a banner made by Ira Deveson referring to ‘no boring bits’.

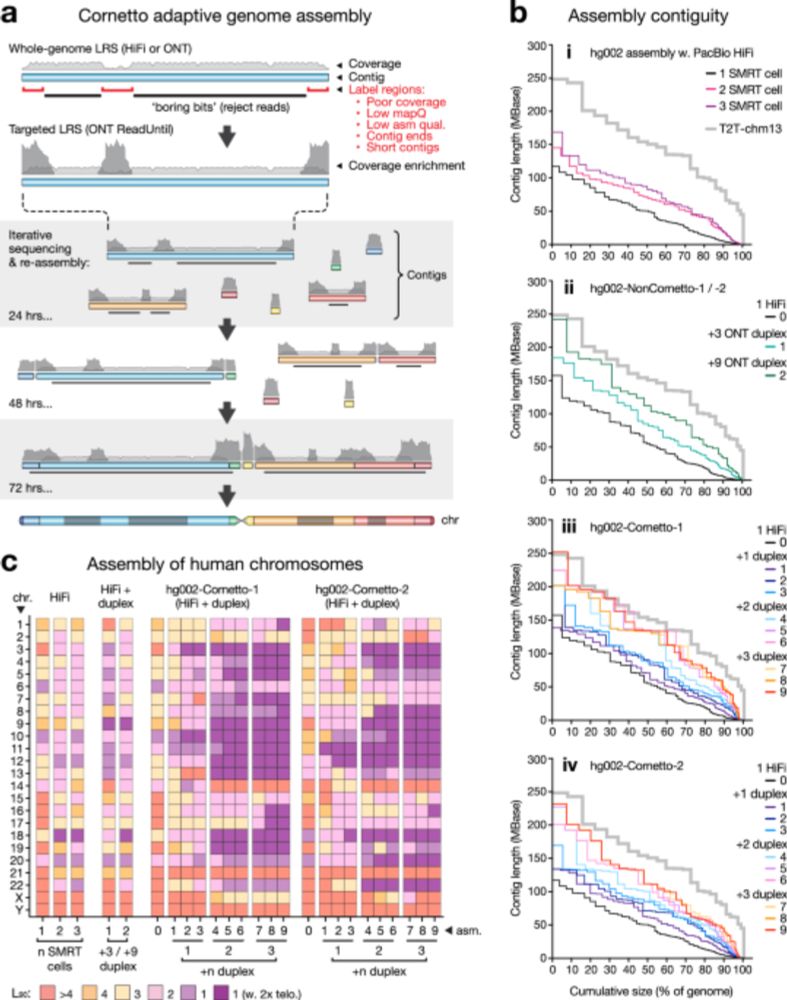
It can do near-T2T assembly using @nanoporetech.com adaptive sampling
- with less 💸
- reference agnostic, so works for non-humans
- not just blood, even saliva
Just presented at #abacbs2025 yesterday.
It can do near-T2T assembly using @nanoporetech.com adaptive sampling
- with less 💸
- reference agnostic, so works for non-humans
- not just blood, even saliva
Just presented at #abacbs2025 yesterday.
PhD candidate @bonson-wong.bsky.social (poster at #abacbs2025) and great to see being highlighted in the AMD blog: www.amd.com/en/blogs/202...
PhD candidate @bonson-wong.bsky.social (poster at #abacbs2025) and great to see being highlighted in the AMD blog: www.amd.com/en/blogs/202...
@GigaScience
academic.oup.com/gigascience/....
Some plots required a log scale - RAM usage and random access time.

@GigaScience
academic.oup.com/gigascience/....
Some plots required a log scale - RAM usage and random access time.
Reads lengths: 9.0 kbases median, 17.2 kbases mean
See:
gentechgp.github.io/gtgseq/docs/...
Reads lengths: 9.0 kbases median, 17.2 kbases mean
See:
gentechgp.github.io/gtgseq/docs/...
github.com/hasindu2008/...
If you have any AMD GPUs lying around, help us test the binaries.

github.com/hasindu2008/...
If you have any AMD GPUs lying around, help us test the binaries.
github.com/hasindu2008/...
Cornetto is not only an iterative assembly method for @nanoporetech.com, but also features some basic assembly evaluation tools that we implemented.
Suggestions and feedback welcome!

github.com/hasindu2008/...
Cornetto is not only an iterative assembly method for @nanoporetech.com, but also features some basic assembly evaluation tools that we implemented.
Suggestions and feedback welcome!
Nanopore's getting accurate, but
1. Can this lead to better metagenome assemblies?
2. How, algorithmically, to leverage them?
with co-author Max Marin @mgmarin.bsky.social, supervised by Heng Li @lh3lh3.bsky.social
1 / N
Nanopore's getting accurate, but
1. Can this lead to better metagenome assemblies?
2. How, algorithmically, to leverage them?
with co-author Max Marin @mgmarin.bsky.social, supervised by Heng Li @lh3lh3.bsky.social
1 / N
Learn more: youtu.be/FL8ZKUmtYbM
#EurekaPrizes

biorxiv.org/content/10.1...
-similar accuracy to modkit & pb-CpG-tools.
-standard open-source licenses (NOT vendor-specific)
-Simple but faster, on a laptop ~4X for DNA and ~55X for RNA.
Code: github.com/warp9seq/min...
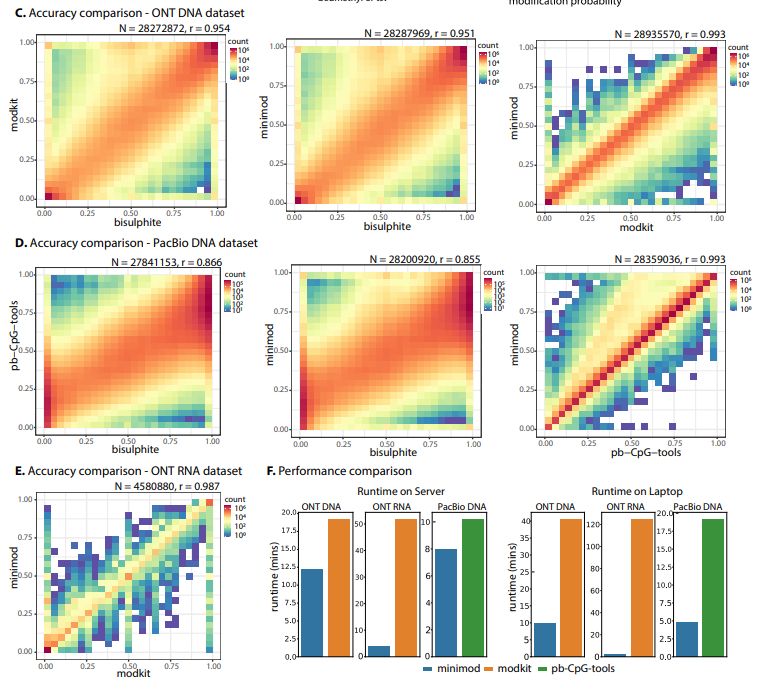
biorxiv.org/content/10.1...
-similar accuracy to modkit & pb-CpG-tools.
-standard open-source licenses (NOT vendor-specific)
-Simple but faster, on a laptop ~4X for DNA and ~55X for RNA.
Code: github.com/warp9seq/min...
@bosc.bsky.social track around 2:30pm ish after
@sunethsa.bsky.social's talk, Bonson Wong will present on
nanopore basecalling on AMD GPUs using slorado
github.com/BonsonW/slor...
@bosc.bsky.social track around 2:30pm ish after
@sunethsa.bsky.social's talk, Bonson Wong will present on
nanopore basecalling on AMD GPUs using slorado
github.com/BonsonW/slor...
@bosc.bsky.social around 2:30pm ish where
@sunethsa.bsky.social will present real-time @nanoporetech.com frequency calculation using realfreq & standalone frequency calculation using minimod.
academic.oup.com/bioinformati...
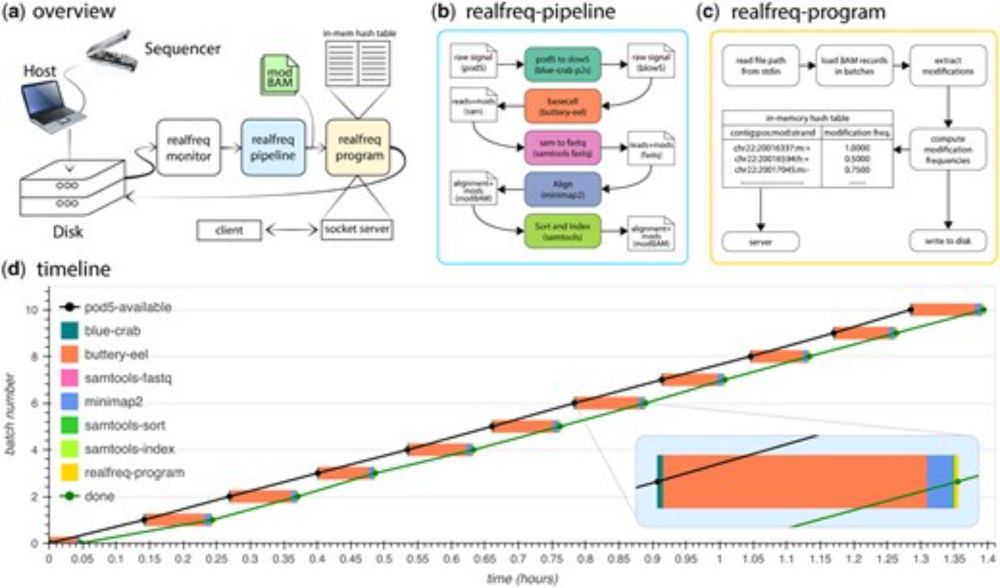
@bosc.bsky.social around 2:30pm ish where
@sunethsa.bsky.social will present real-time @nanoporetech.com frequency calculation using realfreq & standalone frequency calculation using minimod.
academic.oup.com/bioinformati...
github.com/warp9seq/min...
written by
@sunethsa.bsky.social
in C, based on mod tag parsing we did for realfreq doi.org/10.1093/bioi...
github.com/warp9seq/min...
written by
@sunethsa.bsky.social
in C, based on mod tag parsing we did for realfreq doi.org/10.1093/bioi...
- yet another end_reason added to support pod5 updates.
To convert POD5<=>S/BLOW5 it's as simple as
pip install blue-crab
pod5->blow5
blue-crab p2s example.pod5 -o example.blow5
blow5->pod5
blue-crab s2p example.blow5 -o example.pod5
github.com/Psy-Fer/blue...

- yet another end_reason added to support pod5 updates.
To convert POD5<=>S/BLOW5 it's as simple as
pip install blue-crab
pod5->blow5
blue-crab p2s example.pod5 -o example.blow5
blow5->pod5
blue-crab s2p example.blow5 -o example.pod5
github.com/Psy-Fer/blue...
biorxiv.org/content/10.1...
Summary: performance of BLOW5 is >= POD5 (from ~= to 100X, see below), with benefit of having ~3 dependencies instead of >50.
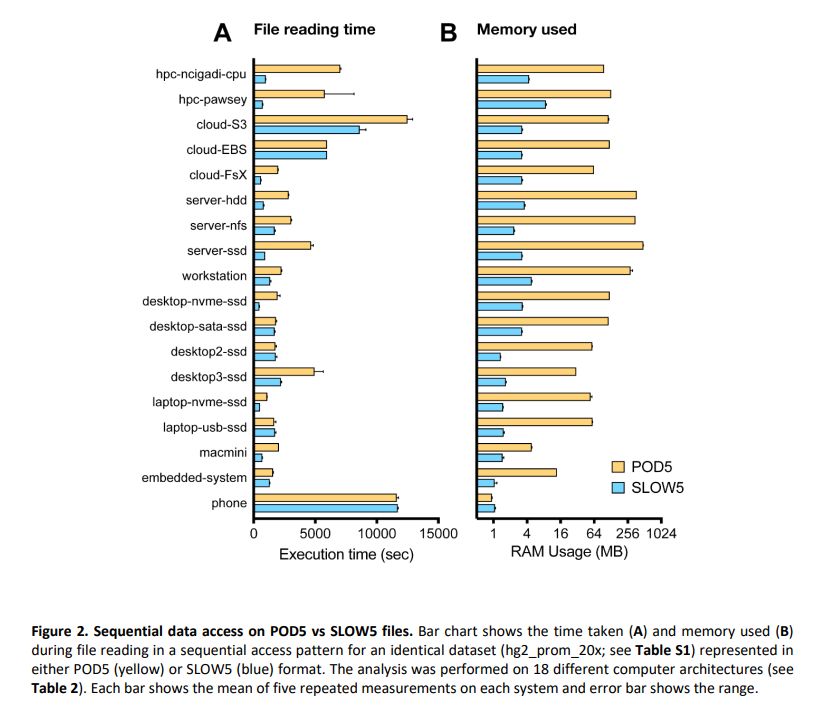
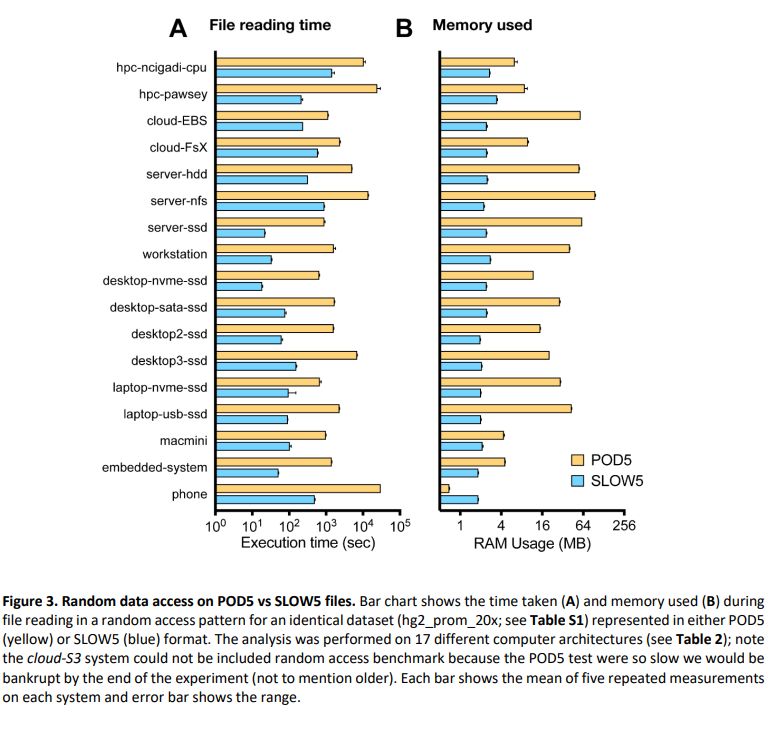
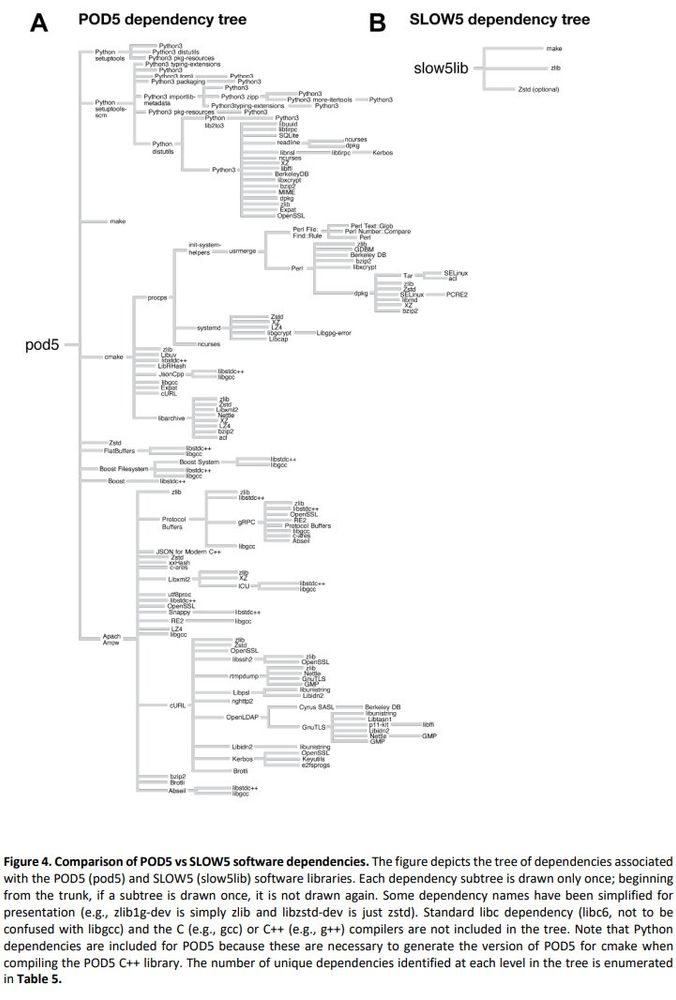
biorxiv.org/content/10.1...
Summary: performance of BLOW5 is >= POD5 (from ~= to 100X, see below), with benefit of having ~3 dependencies instead of >50.
genome.cshlp.org/content/35/7...

genome.cshlp.org/content/35/7...
www.biorxiv.org/content/10.1...
(1/6)

www.biorxiv.org/content/10.1...
(1/6)

On May 27th at 10 AM (CEST), @hasindu2008.bsky.social will give a talk on "Scalable, Efficient and Real-time Analysis of Long-read Genomic Data"
More on this talk: safari.ethz.ch/safari-efcl-... (1/3)
On May 27th at 10 AM (CEST), @hasindu2008.bsky.social will give a talk on "Scalable, Efficient and Real-time Analysis of Long-read Genomic Data"
More on this talk: safari.ethz.ch/safari-efcl-... (1/3)
Written efficiently using C, a laptop can keep up with a
@nanoporetech.com MinION sequencer, and a desktop a PromethION 2 solo flowcell.
academic.oup.com/bioinformati...

Written efficiently using C, a laptop can keep up with a
@nanoporetech.com MinION sequencer, and a desktop a PromethION 2 solo flowcell.
academic.oup.com/bioinformati...

