Designing embedded systems for bioinformatics applications.

@GigaScience
academic.oup.com/gigascience/....
Some plots required a log scale - RAM usage and random access time.

@GigaScience
academic.oup.com/gigascience/....
Some plots required a log scale - RAM usage and random access time.
It is very interesting that they have skipped those questions 🤣

It is very interesting that they have skipped those questions 🤣
biorxiv.org/content/10.1...
-similar accuracy to modkit & pb-CpG-tools.
-standard open-source licenses (NOT vendor-specific)
-Simple but faster, on a laptop ~4X for DNA and ~55X for RNA.
Code: github.com/warp9seq/min...
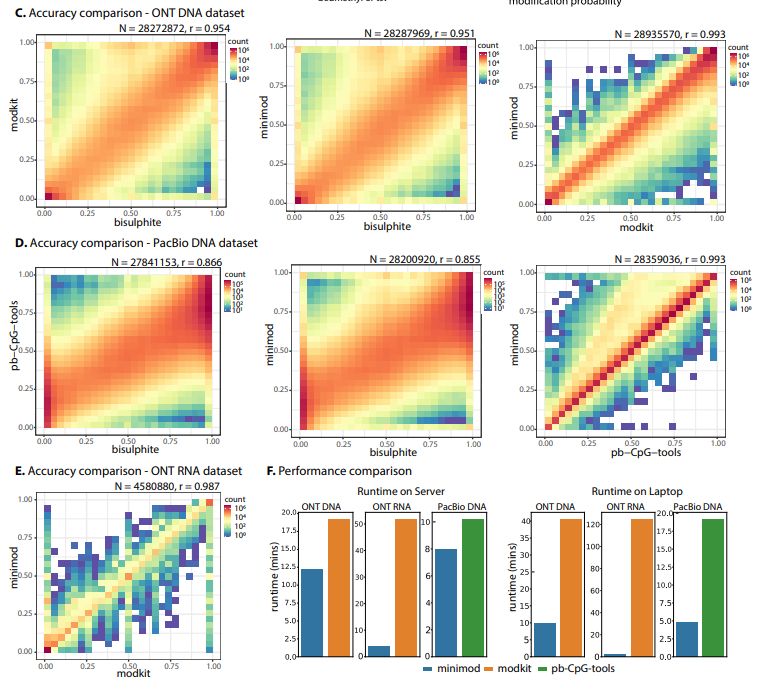
biorxiv.org/content/10.1...
-similar accuracy to modkit & pb-CpG-tools.
-standard open-source licenses (NOT vendor-specific)
-Simple but faster, on a laptop ~4X for DNA and ~55X for RNA.
Code: github.com/warp9seq/min...
6754
6751
6769
6760
6756
Which seem to match the expected, assuming you are using 1-based coordinates
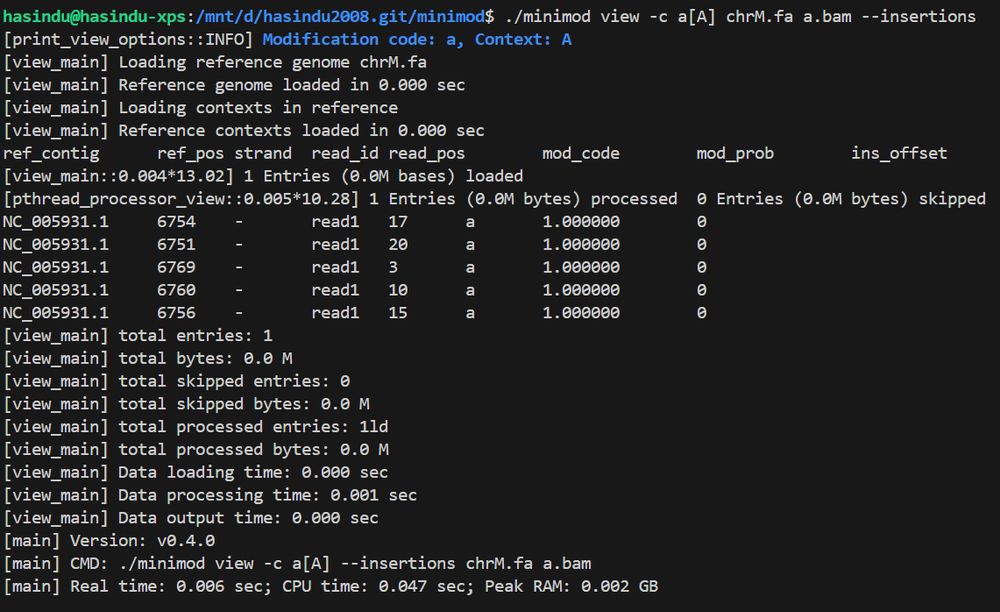
6754
6751
6769
6760
6756
Which seem to match the expected, assuming you are using 1-based coordinates
biorxiv.org/content/10.1...
Summary: performance of BLOW5 is >= POD5 (from ~= to 100X, see below), with benefit of having ~3 dependencies instead of >50.
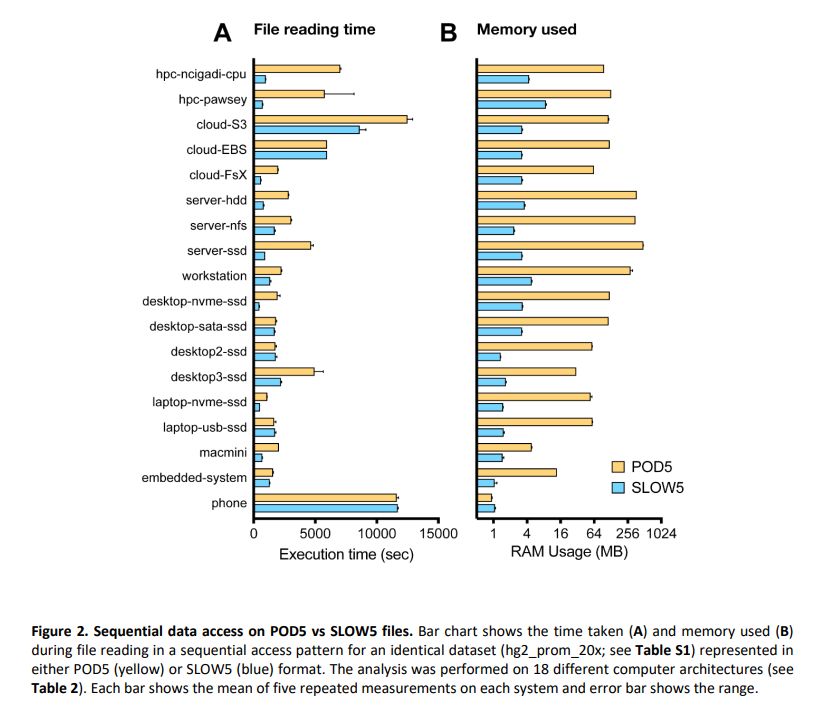
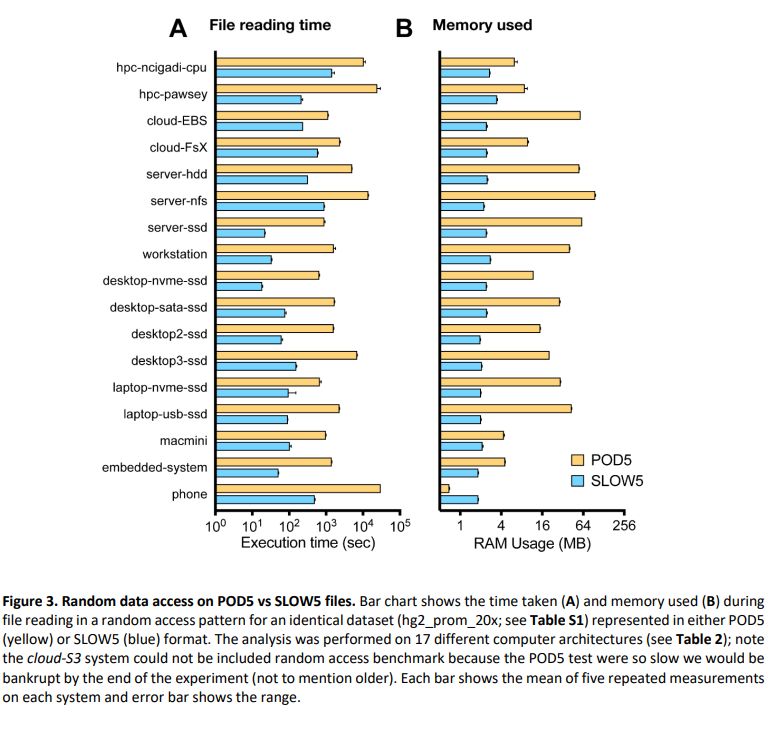
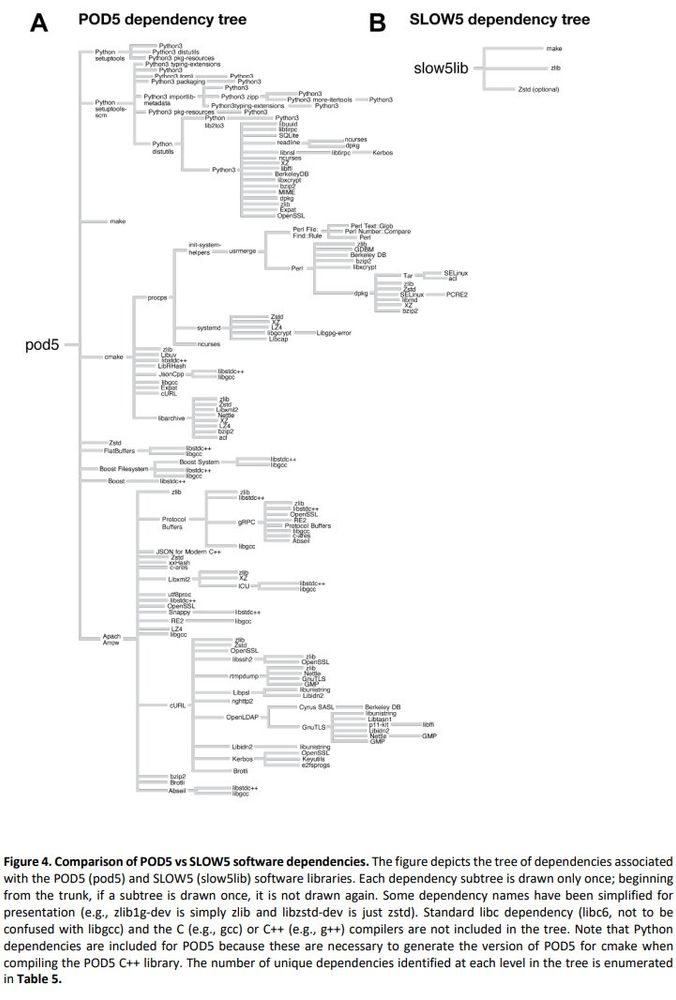
biorxiv.org/content/10.1...
Summary: performance of BLOW5 is >= POD5 (from ~= to 100X, see below), with benefit of having ~3 dependencies instead of >50.
genome.cshlp.org/content/35/7...

genome.cshlp.org/content/35/7...


