www.science.org/doi/10.1126/...
🧵

www.science.org/doi/10.1126/...
🧵

Let us take a tour through proteotoxic stress in intact human tissue — one hepatocyte at a time.
www.nature.com/articles/s41...
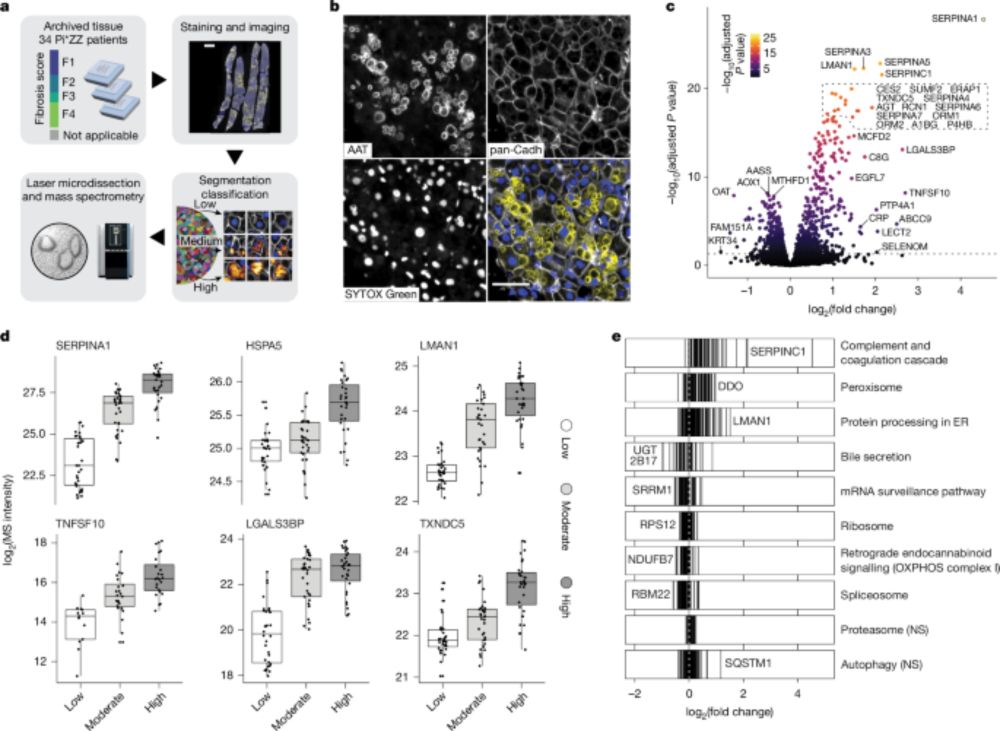
Let us take a tour through proteotoxic stress in intact human tissue — one hepatocyte at a time.
www.nature.com/articles/s41...


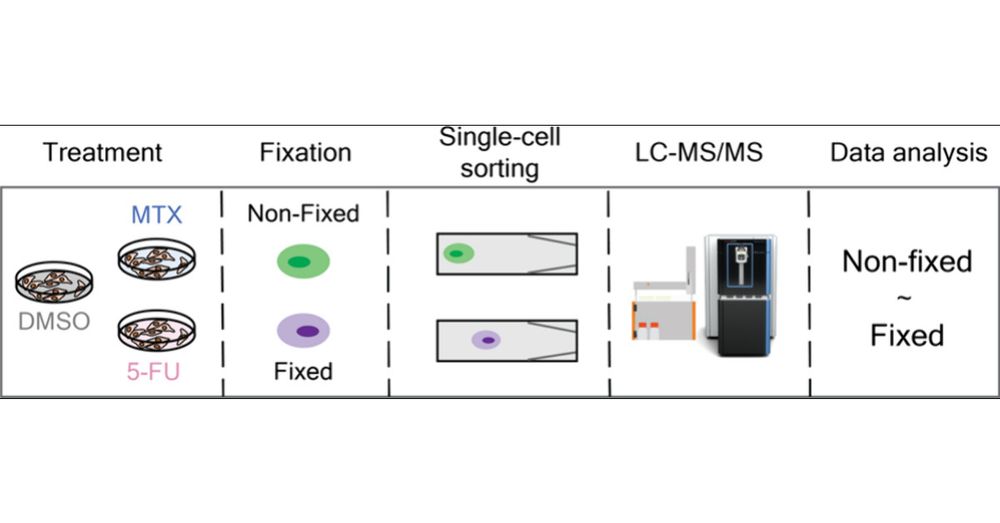
We evaluated how formaldehyde-based fixation preserve proteome state, drug response and cell integrity. Ultimately, cell fixation can facilitate access to #singlecell by enabeling sample shiping and prolonged sorting 📦
pubs.acs.org/doi/10.1021/...
pubs.acs.org/doi/10.1021/...
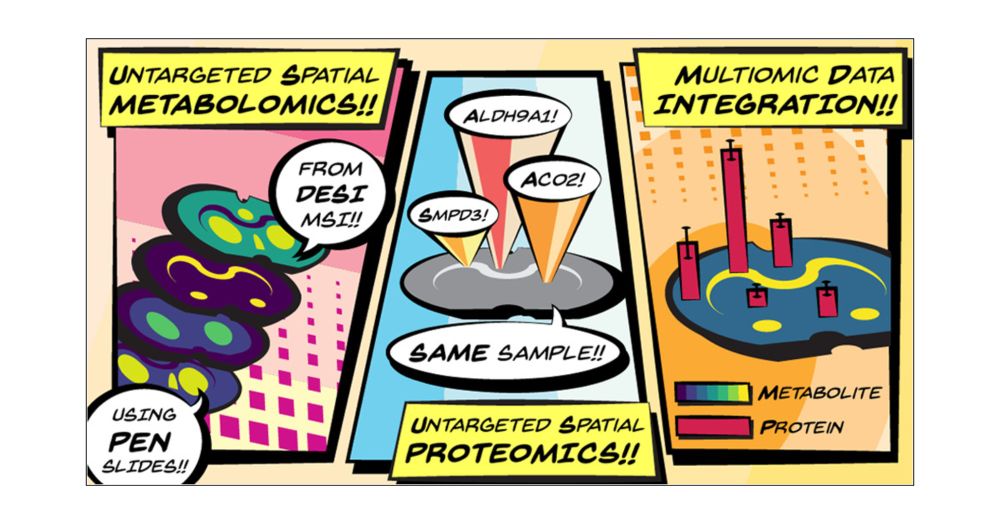

www.nature.com/articles/s41...

We would be delighted to receive many abstracts from students so that they have the opportunity to give their first presentation.
On behalf of: Erwin, Fabian, Manuel, Fabian and Karl
We would be delighted to receive many abstracts from students so that they have the opportunity to give their first presentation.
On behalf of: Erwin, Fabian, Manuel, Fabian and Karl

