
Lab website: http://transdevolab.com
www.evomg-dn.eu/project/mole...

www.evomg-dn.eu/project/mole...
www.evomg-dn.eu/project/evol...

www.evomg-dn.eu/project/evol...
www.annualreviews.org/content/jour...
www.annualreviews.org/content/jour...
Evolution of comparative transcriptomics: biological scales, phylogenetic spans, and modeling frameworks
authors.elsevier.com/sd/article/S...
By @mattezambon.bsky.social & @fedemantica.bsky.social, together with @jonnyfrazer.bsky.social & Mafalda Dias.
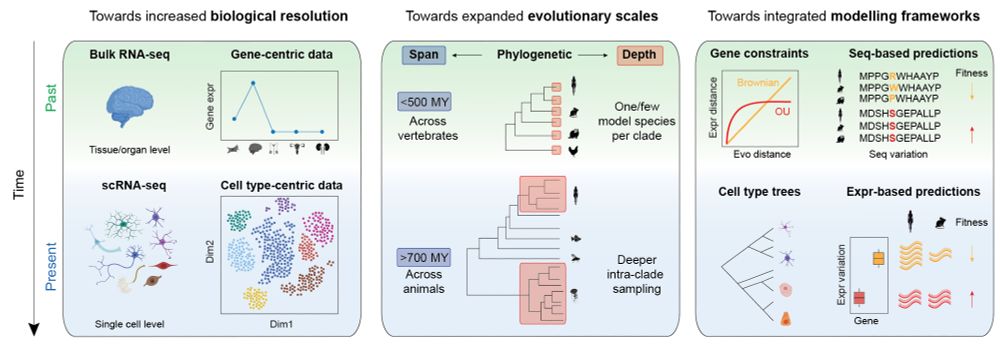
Evolution of comparative transcriptomics: biological scales, phylogenetic spans, and modeling frameworks
authors.elsevier.com/sd/article/S...
By @mattezambon.bsky.social & @fedemantica.bsky.social, together with @jonnyfrazer.bsky.social & Mafalda Dias.

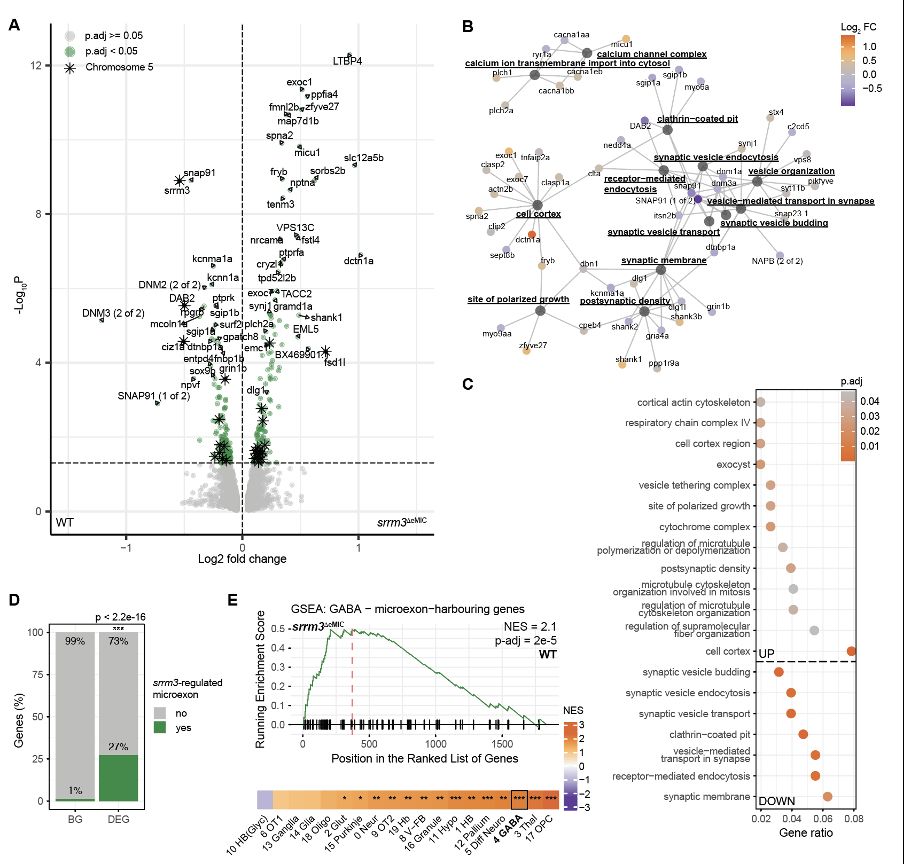
● Inhibiting adenylyl cyclases (reducing cAMP) rescued mutant hyperactivity.
● Increasing cAMP in the wild-type phenocopied the mutant.
8/9

● Inhibiting adenylyl cyclases (reducing cAMP) rescued mutant hyperactivity.
● Increasing cAMP in the wild-type phenocopied the mutant.
8/9




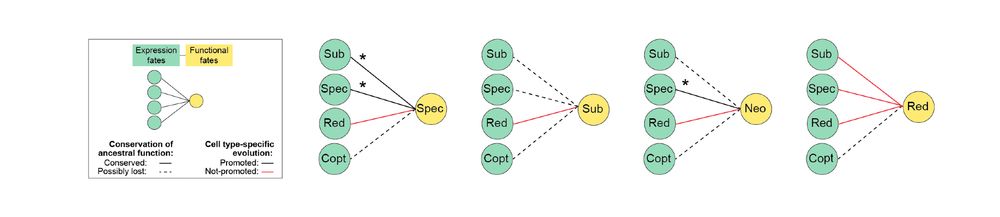

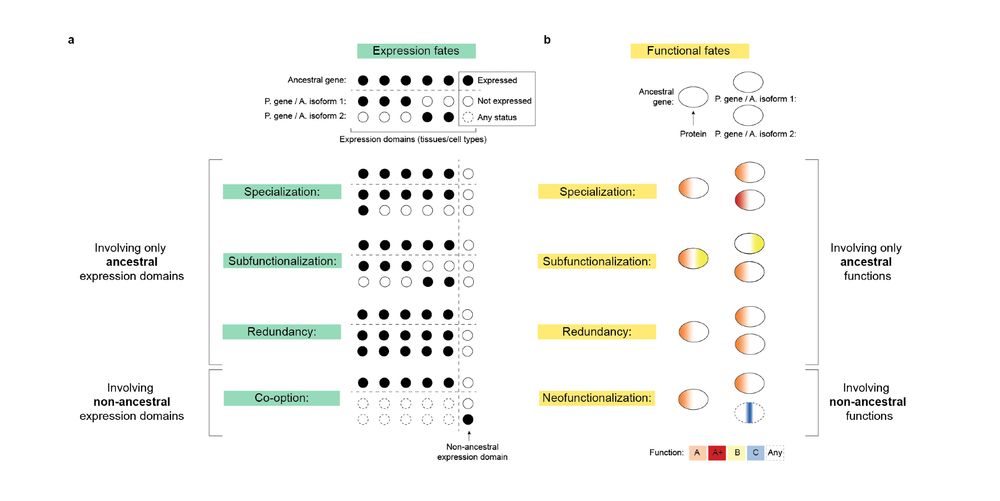


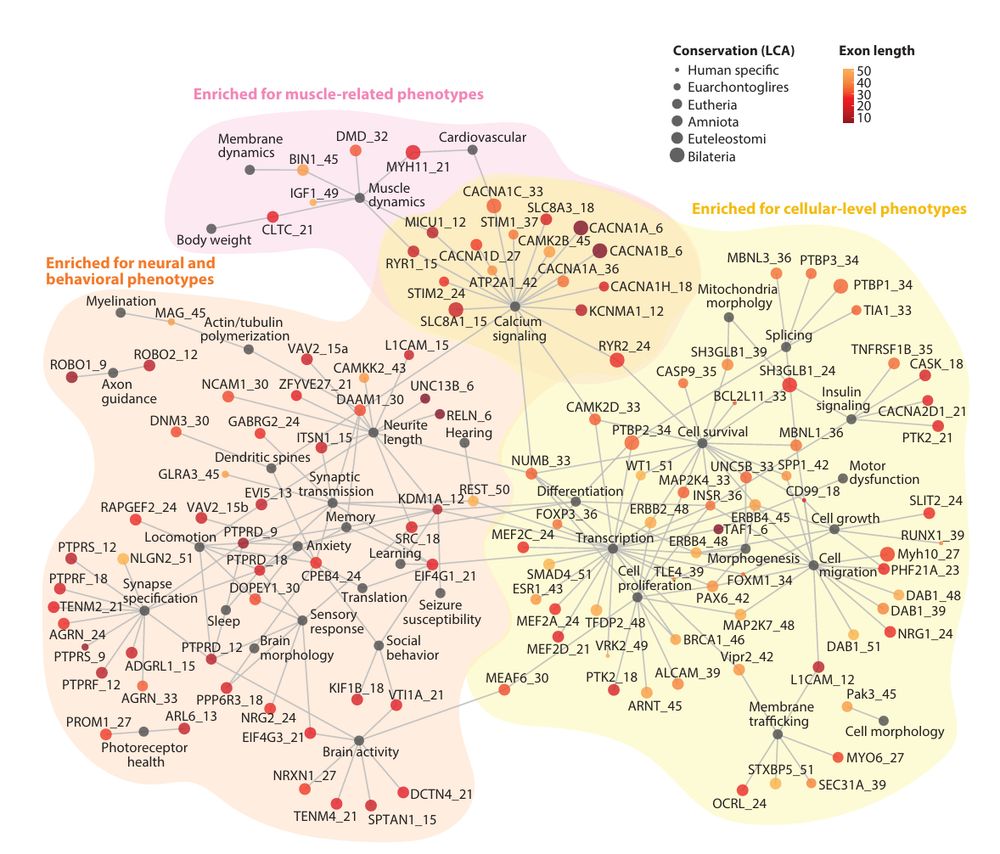
In our new review, @fedemantica.bsky.social and I argue we are missing the most prevalent one: specialization. And the same applies to alternative splicing! 1/7
tinyurl.com/45k7kbmp
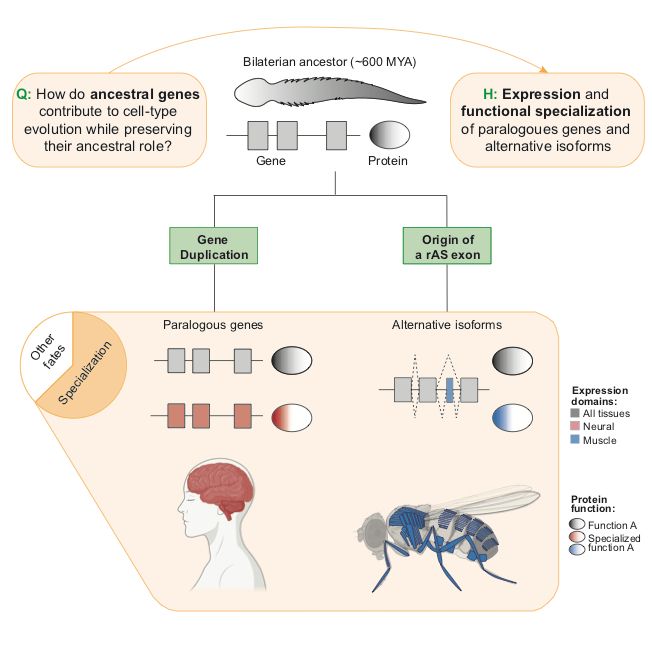
In our new review, @fedemantica.bsky.social and I argue we are missing the most prevalent one: specialization. And the same applies to alternative splicing! 1/7
tinyurl.com/45k7kbmp

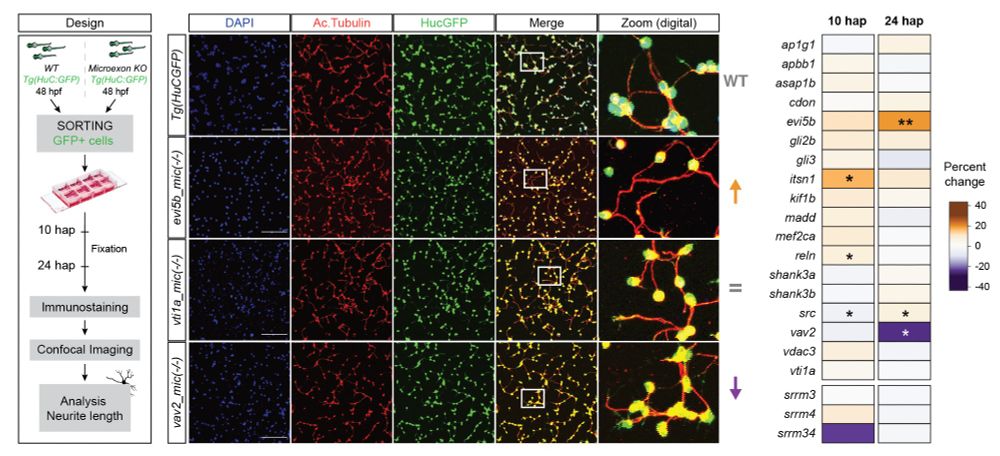



If you're a bioinformatician who loves:
🧬 #SingleCell #LongReadSequencing & #AlternativeSplicing
🚀 Collaborating and travelling
Apply to work with us at the spectacular BIMSB (@mdc-berlin.bsky.social) in Berlin + @upf.edu-@crg.eu in Barcelona. Qs: www.transdevolab.com
Please RT! 🙏

If you're a bioinformatician who loves:
🧬 #SingleCell #LongReadSequencing & #AlternativeSplicing
🚀 Collaborating and travelling
Apply to work with us at the spectacular BIMSB (@mdc-berlin.bsky.social) in Berlin + @upf.edu-@crg.eu in Barcelona. Qs: www.transdevolab.com
Please RT! 🙏
www.crg.eu/en/content/t...

www.crg.eu/en/content/t...

