
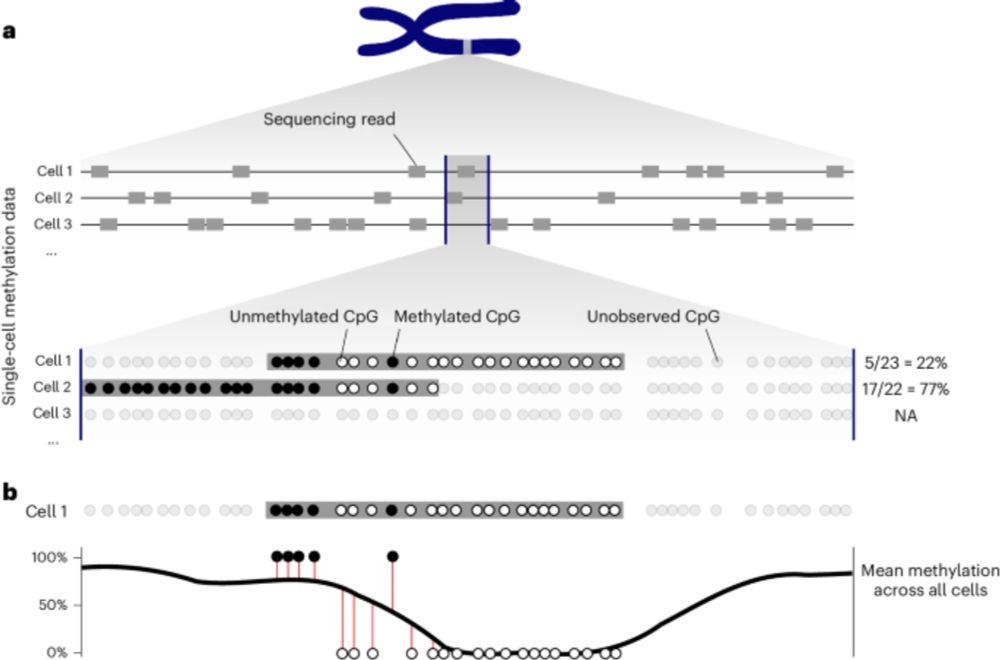
🧬 Single cell methylome encodes cell state & clonal identity
🔨 EPI-Clone reads out both (+mutations, +RNA) at scale
🩸 Clonal expansions of HSCs are universal from age 50, not driven by CH mutations
doi.org/10.1038/s415...
🧵
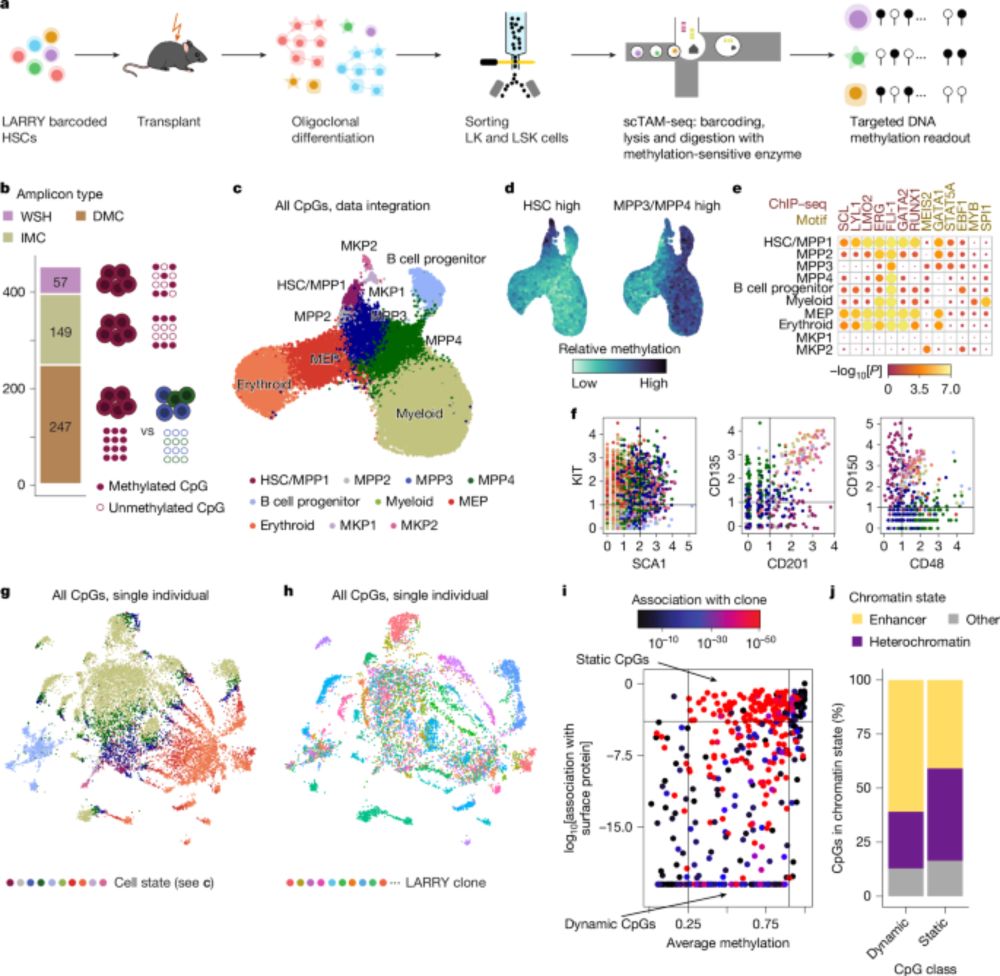
I loved analyzing the human data - we discovered that after age 50, blood production shifts to an oligoclonal pattern, and these expansions aren’t driven by CH mutations!
Dive into @larsplus.bsky.social thread for the full story & visuals.
genzplyr is dplyr, but bussin fr fr no cap.

genzplyr is dplyr, but bussin fr fr no cap.
@karalmckinley.bsky.social
We built the first transgenic model of menstruation in mice.
We used it to uncover how the endometrium organizes and sheds during menstruation. 🧪
www.biorxiv.org/content/10.1...
🧵

@karalmckinley.bsky.social
We built the first transgenic model of menstruation in mice.
We used it to uncover how the endometrium organizes and sheds during menstruation. 🧪
www.biorxiv.org/content/10.1...
🧵
Very happy to share the results of a great collaboration between @hovestadt.bsky.social and Griffin Labs, I co-led with @sbenfatto.bsky.social, published today in @natgenet.nature.com
📄 www.nature.com/articles/s41...
🧵1/n

Very happy to share the results of a great collaboration between @hovestadt.bsky.social and Griffin Labs, I co-led with @sbenfatto.bsky.social, published today in @natgenet.nature.com
📄 www.nature.com/articles/s41...
🧵1/n
🔗 Manuscript: www.biorxiv.org/content/10.1...
💻 Code: partipy.readthedocs.io

🔗 Manuscript: www.biorxiv.org/content/10.1...
💻 Code: partipy.readthedocs.io

👉 Early cancer detection
👉 Methylation drivers
👉 Epimutation rates
👉 CpG lineage tracing
www.biorxiv.org/content/10.1...
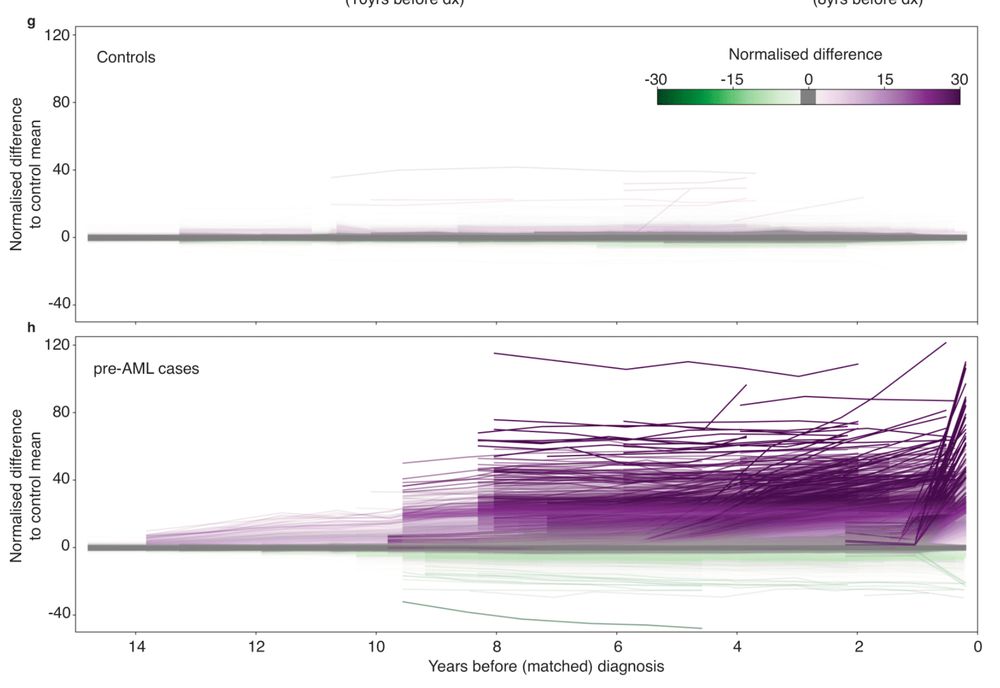
👉 Early cancer detection
👉 Methylation drivers
👉 Epimutation rates
👉 CpG lineage tracing
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
https://go.nature.com/43NOe1R
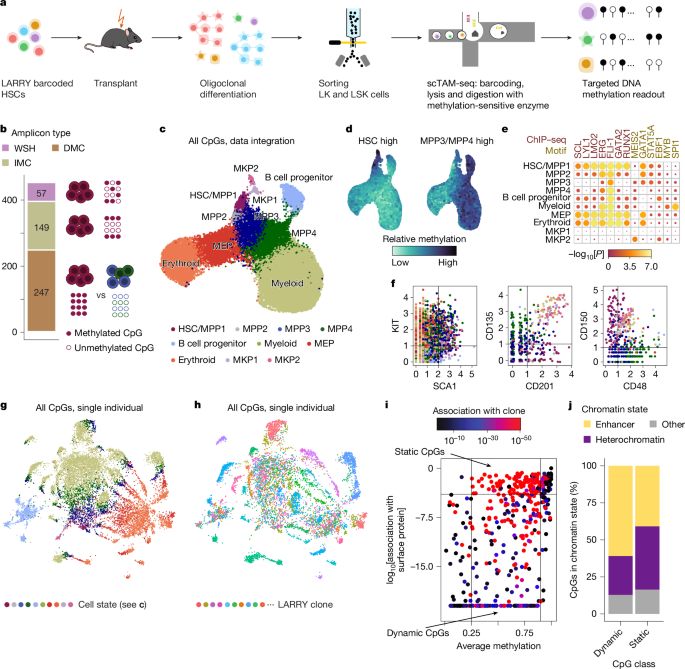
https://go.nature.com/43NOe1R
Check out the thread by @larsplus.bsky.social below.
EPI-clone is a revolutionary technique for lineage tracing and it will immediately impact how we trace clones everywhere.
Detailed protocols coming out soon!
🧬 Single cell methylome encodes cell state & clonal identity
🔨 EPI-Clone reads out both (+mutations, +RNA) at scale
🩸 Clonal expansions of HSCs are universal from age 50, not driven by CH mutations
doi.org/10.1038/s415...
🧵

Check out the thread by @larsplus.bsky.social below.
EPI-clone is a revolutionary technique for lineage tracing and it will immediately impact how we trace clones everywhere.
Detailed protocols coming out soon!
I loved analyzing the human data - we discovered that after age 50, blood production shifts to an oligoclonal pattern, and these expansions aren’t driven by CH mutations!
Dive into @larsplus.bsky.social thread for the full story & visuals.
🧬 Single cell methylome encodes cell state & clonal identity
🔨 EPI-Clone reads out both (+mutations, +RNA) at scale
🩸 Clonal expansions of HSCs are universal from age 50, not driven by CH mutations
doi.org/10.1038/s415...
🧵

I loved analyzing the human data - we discovered that after age 50, blood production shifts to an oligoclonal pattern, and these expansions aren’t driven by CH mutations!
Dive into @larsplus.bsky.social thread for the full story & visuals.
🧬 Single cell methylome encodes cell state & clonal identity
🔨 EPI-Clone reads out both (+mutations, +RNA) at scale
🩸 Clonal expansions of HSCs are universal from age 50, not driven by CH mutations
doi.org/10.1038/s415...
🧵

Naturally occurring methylation patterns reveal changes in blood which are detectable at age 50 and almost universal by 60
📰 Published in @nature.com by #IRBBarcelona & @crg.eu
Read the news ➡️ bit.ly/4jZHQeB
⬇️ ⬇️
Naturally occurring methylation patterns reveal changes in blood which are detectable at age 50 and almost universal by 60
📰 Published in @nature.com by #IRBBarcelona & @crg.eu
Read the news ➡️ bit.ly/4jZHQeB
⬇️ ⬇️
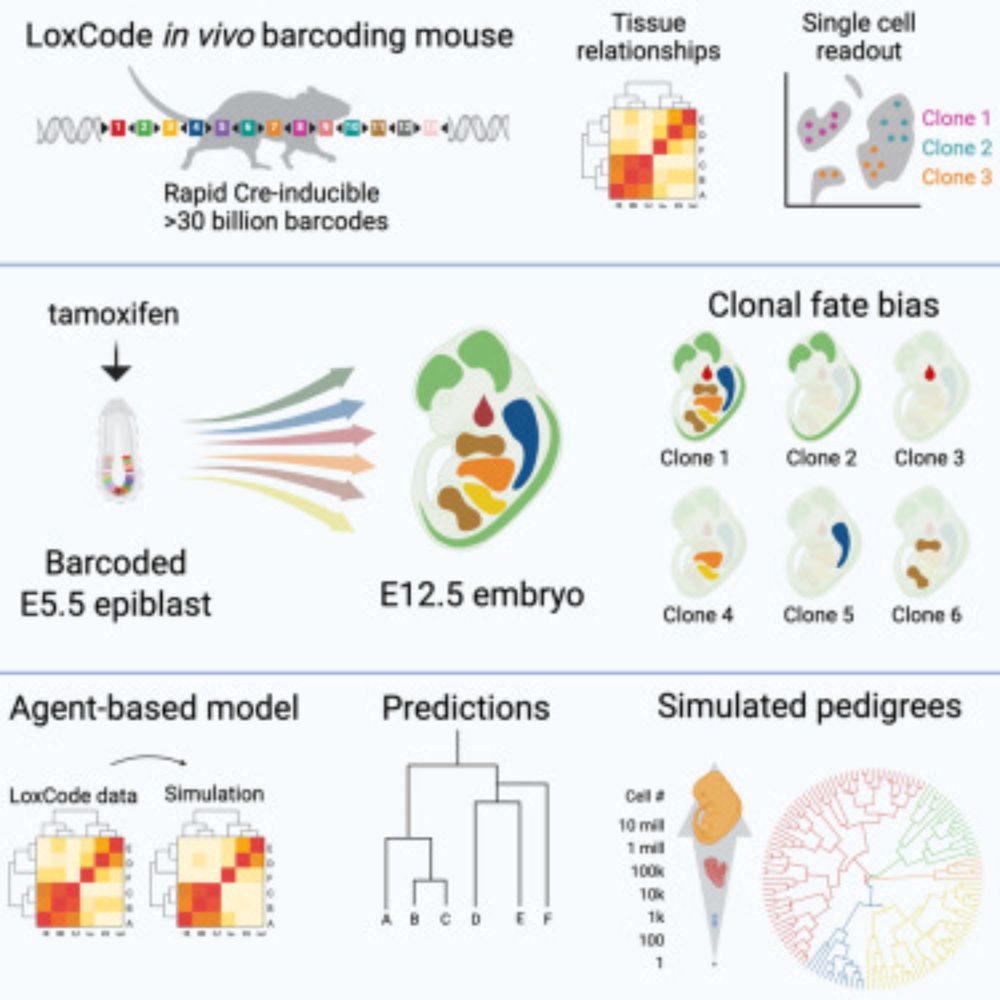
Check out our preprint for more. Full paper coming soon! www.biorxiv.org/content/10.1...

Check out our preprint for more. Full paper coming soon! www.biorxiv.org/content/10.1...
I'm so excited about this!
www.biorxiv.org/content/10.1...

I'm so excited about this!
www.biorxiv.org/content/10.1...
🧬🩸 screen of fully synthetic enhancers in blood progenitors
🤖 AI that creates new cell state specific enhancers
🔍 negative synergies between TFs lead to specificity!
www.cell.com/cell/fulltex...
🧵

“Pre-existing stem cell heterogeneity dictates clonal responses to the acquisition of leukemia driver mutations”
Now at Cell Stem Cell, with two new figures, in vivo, and sequential mutagenesis data.
Performed with the support of Cris Cancer and @erc.europa.eu
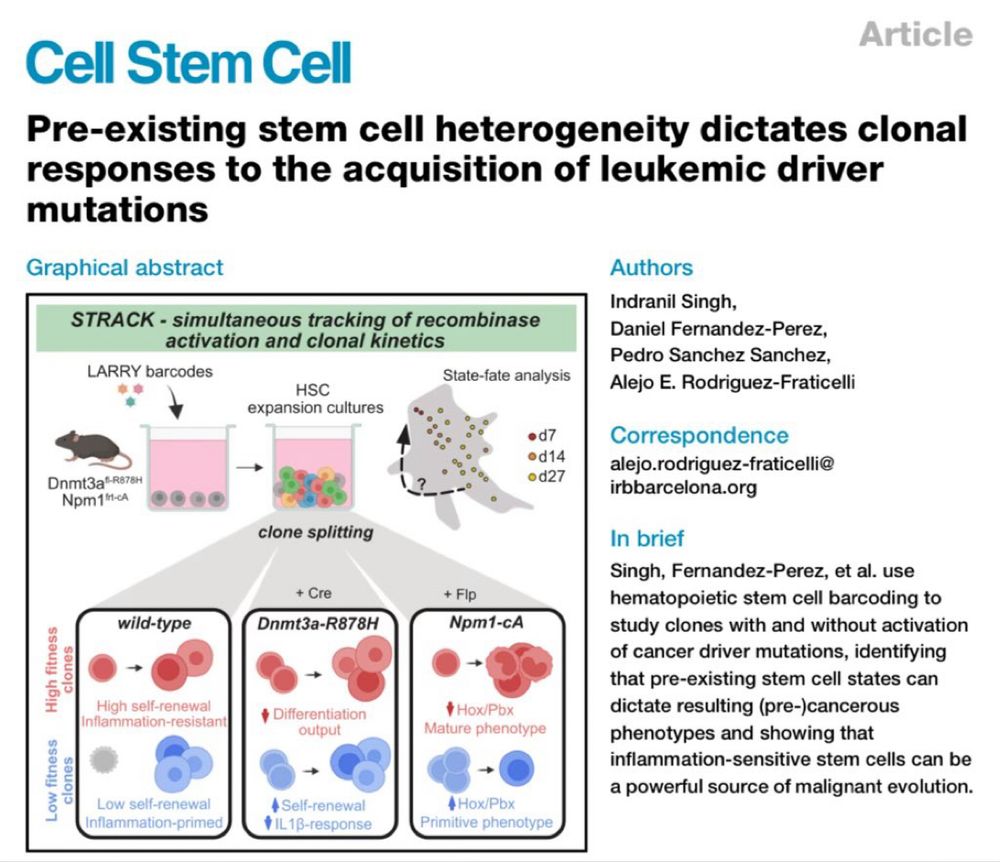
“Pre-existing stem cell heterogeneity dictates clonal responses to the acquisition of leukemia driver mutations”
Now at Cell Stem Cell, with two new figures, in vivo, and sequential mutagenesis data.
Performed with the support of Cris Cancer and @erc.europa.eu
www.nature.com/articles/s41...

www.nature.com/articles/s41...
Looking forward to sharing more thoughts with the community. 🧬✨🚀
www.nature.com/articles/s41...
Looking forward to sharing more thoughts with the community. 🧬✨🚀

