Mark Polk
@markpolk.io
I use robots and biophysics to study cancer therapies in the Chodera Lab at Memorial Sloan Kettering Cancer Center. Views are my own.
Pinned
Mark Polk
@markpolk.io
· Nov 27
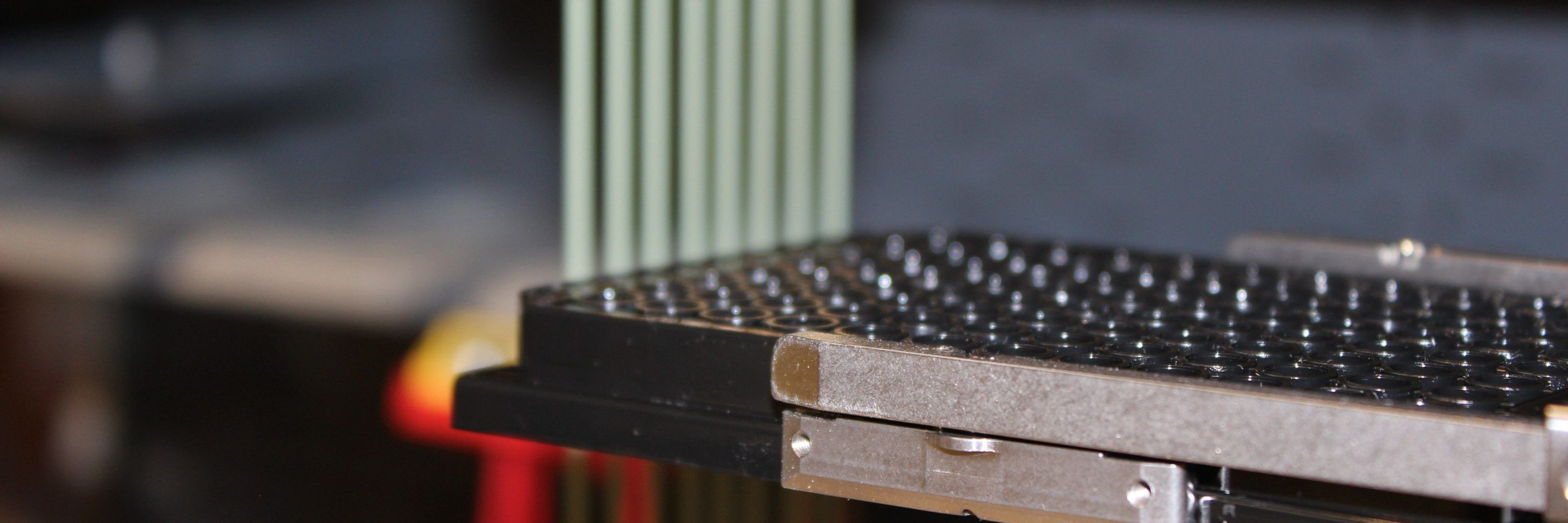
Automated plate handling in the Chodera Lab, part of our effort to study kinase inhibitors in high throughput.
Reposted by Mark Polk
How many crystal structures do you need to trust your docking results? https://www.biorxiv.org/content/10.1101/2025.09.19.677428v1
September 25, 2025 at 2:48 AM
How many crystal structures do you need to trust your docking results? https://www.biorxiv.org/content/10.1101/2025.09.19.677428v1
Reposted by Mark Polk
Join us for the NYAGIM 2025 Q3 event, featuring Dan Sindhikara on recent advances and persistent challenges in the computational design of macrocyclic peptides. Sep. 16 at 6 PM. RSVP: www.meetup.com/new-york-are...

Computational Design of Macrocyclic Peptides Containing Non-Natural Amino Acids , Tue, Sep 16, 2025, 6:00 PM | Meetup
Join us for the NYAGIM 2025 Q3 event, supported by Cresset and the New York Academy of Sciences, featuring Dan Sindhikara on recent advances and persistent challenges in th
www.meetup.com
September 4, 2025 at 3:20 PM
Join us for the NYAGIM 2025 Q3 event, featuring Dan Sindhikara on recent advances and persistent challenges in the computational design of macrocyclic peptides. Sep. 16 at 6 PM. RSVP: www.meetup.com/new-york-are...
In Edinburgh for the RSC/SCI Symposium on Kinase Inhibitor Design—excited to see @jessbwhite.bsky.social share her work on the missense kinase toolkit! #Kinase2025

May 13, 2025 at 10:52 AM
In Edinburgh for the RSC/SCI Symposium on Kinase Inhibitor Design—excited to see @jessbwhite.bsky.social share her work on the missense kinase toolkit! #Kinase2025
Reposted by Mark Polk
the longest journey begins with a single step. luckily for us, @arianaclerkin.bsky.social is writing hers down!
read the first PLUMB blab of many below, and follow along as Ariana builds an open comp.chem project from the ground up.
read the first PLUMB blab of many below, and follow along as Ariana builds an open comp.chem project from the ground up.
PLUMB blab #1: We need better benchmarks for computer-aided drug design
Written by: Ariana Brenner Clerkin, PhD
Welcome to a new series on the OMSF blog: PLUMB blab! This series will take you behind the scenes of PLUMB (Protein-Ligand Unified Metrics Benchmark), an open-...
omsf-blog.ghost.io
April 1, 2025 at 3:14 PM
the longest journey begins with a single step. luckily for us, @arianaclerkin.bsky.social is writing hers down!
read the first PLUMB blab of many below, and follow along as Ariana builds an open comp.chem project from the ground up.
read the first PLUMB blab of many below, and follow along as Ariana builds an open comp.chem project from the ground up.
Reposted by Mark Polk
Check out our recent preprint 👇on counting particles to estimate populations in #cryoem: noise can bias the estimates; with Luke Evans & a great team.
Counting particles could give wrong probabilities in Cryo-Electron Microscopy https://www.biorxiv.org/content/10.1101/2025.03.27.644168v1
March 30, 2025 at 1:29 PM
Check out our recent preprint 👇on counting particles to estimate populations in #cryoem: noise can bias the estimates; with Luke Evans & a great team.
Reposted by Mark Polk
Happy Holi, from the @jchodera.bsky.social lab!!

March 14, 2025 at 9:51 PM
Happy Holi, from the @jchodera.bsky.social lab!!
Reposted by Mark Polk
This work is now published in J Phys Chem B! Check out our work showing that simulations predict the impact of distal mutations on kinase-inhibitor binding, and our experimental NanoBRET dataset of 94 kinase mutations that provide a benchmark for future methods. Link:
pubs.acs.org/doi/full/10....
pubs.acs.org/doi/full/10....

March 10, 2025 at 6:19 PM
This work is now published in J Phys Chem B! Check out our work showing that simulations predict the impact of distal mutations on kinase-inhibitor binding, and our experimental NanoBRET dataset of 94 kinase mutations that provide a benchmark for future methods. Link:
pubs.acs.org/doi/full/10....
pubs.acs.org/doi/full/10....
Reposted by Mark Polk
"The #undergraduates usually bring samples from the projects they are working on. In this case, the data they obtain from this trip may become the last piece of data they need before submitting their #research for #publication"🥳🥳 @actacrystc.iucr.org @actacryste.iucr.org #crystallography #education

February 19, 2025 at 10:40 PM
"The #undergraduates usually bring samples from the projects they are working on. In this case, the data they obtain from this trip may become the last piece of data they need before submitting their #research for #publication"🥳🥳 @actacrystc.iucr.org @actacryste.iucr.org #crystallography #education
Reposted by Mark Polk
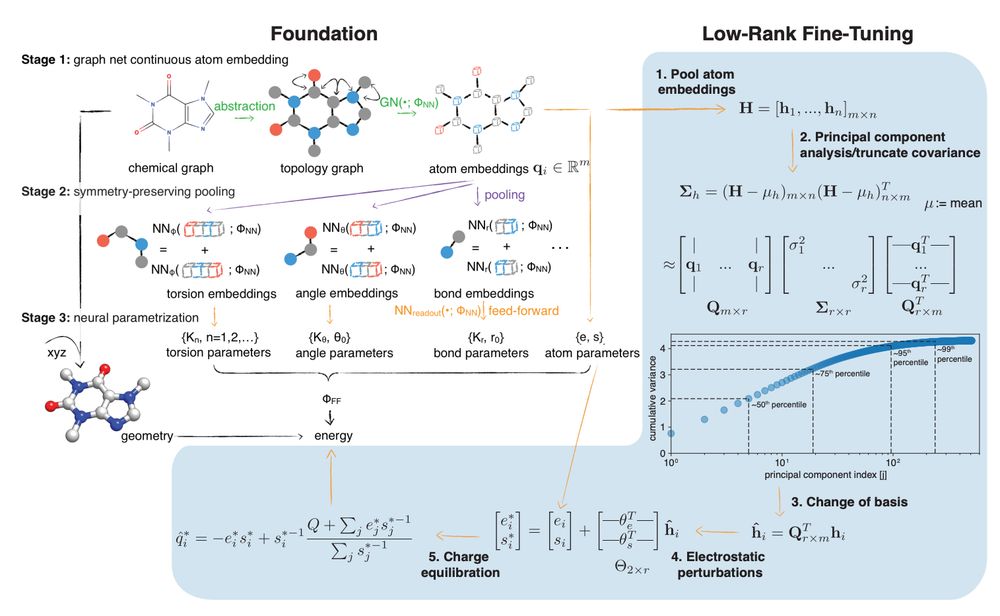
Everything is chaos, but I wanted to share some awesome recent science from the lab that hints at where the future of biomolecular simulation is headed:
Foundation simulation models that can be fine-tuned to experimental free energy data to produce systematically more accurate predictions.
Foundation simulation models that can be fine-tuned to experimental free energy data to produce systematically more accurate predictions.
February 19, 2025 at 7:30 PM
Everything is chaos, but I wanted to share some awesome recent science from the lab that hints at where the future of biomolecular simulation is headed:
Foundation simulation models that can be fine-tuned to experimental free energy data to produce systematically more accurate predictions.
Foundation simulation models that can be fine-tuned to experimental free energy data to produce systematically more accurate predictions.
Reposted by Mark Polk
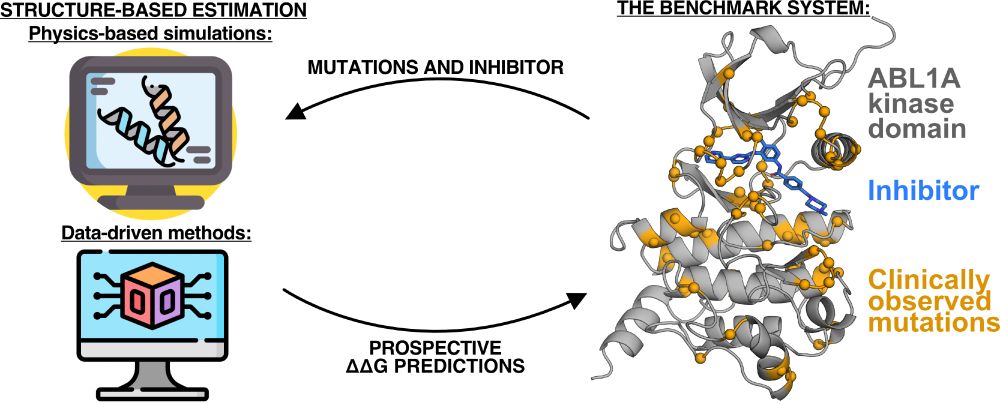
I'll be presenting my work prospectively studying clinical mutations and their impact on kinase inhibitor binding at #bps2025! Come see my talk on Wednesday morning @ 8:45am.
February 16, 2025 at 6:53 PM
I'll be presenting my work prospectively studying clinical mutations and their impact on kinase inhibitor binding at #bps2025! Come see my talk on Wednesday morning @ 8:45am.
Excited to be in LA for #BPS2025 @biophysicalsoc.bsky.social! I’ll be presenting a poster on our high-throughput fluorescence-based assay for kinase inhibitor binding in the West Exhibit Hall on Monday at 2:45 PM (board B60).
February 16, 2025 at 6:47 PM
Excited to be in LA for #BPS2025 @biophysicalsoc.bsky.social! I’ll be presenting a poster on our high-throughput fluorescence-based assay for kinase inhibitor binding in the West Exhibit Hall on Monday at 2:45 PM (board B60).
Reposted by Mark Polk
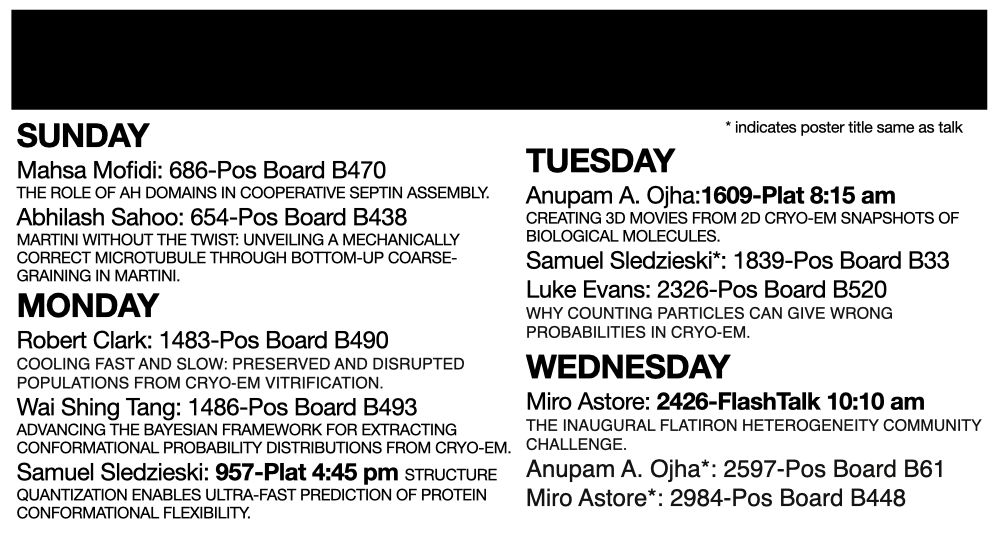
I'm at @biophysicalsoc.bsky.social #BPS2025 with a bunch of folks from the Structural and Molecular Biophysics @flatironinstitute.org group-- come say hi and check out our posters/talks! @sonyahanson.bsky.social @pilarcossio.bsky.social @miroastore.bsky.social
February 15, 2025 at 9:54 PM
I'm at @biophysicalsoc.bsky.social #BPS2025 with a bunch of folks from the Structural and Molecular Biophysics @flatironinstitute.org group-- come say hi and check out our posters/talks! @sonyahanson.bsky.social @pilarcossio.bsky.social @miroastore.bsky.social
Reposted by Mark Polk
Happy to have been a member of the BATCHIE team who worked on this recent publication featured by @MSKLibrary: A Bayesian active learning platform for scalable combination drug screens dx.doi.org/10.1038/s414...
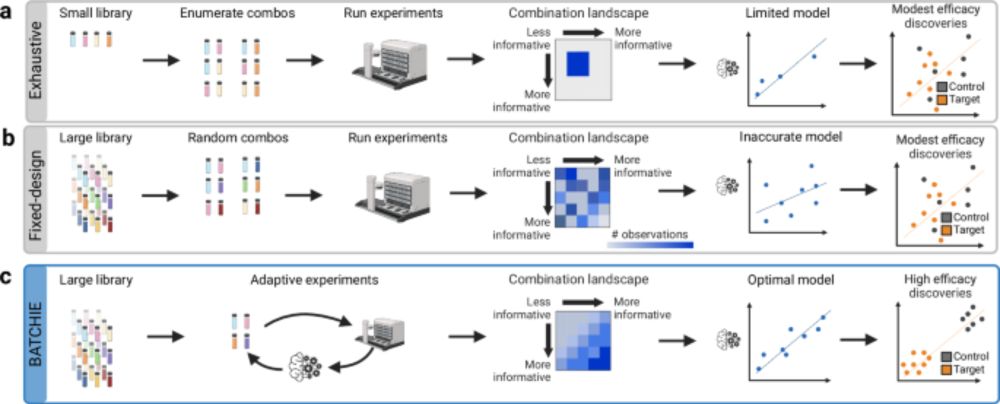
A Bayesian active learning platform for scalable combination drug screens - Nature Communications
Large-scale combination drug screens in cancer are extremely challenging because of the immense number of possible combinations. Here, the authors develop BATCHIE, a Bayesian active learning platform ...
dx.doi.org
January 14, 2025 at 3:04 PM
Happy to have been a member of the BATCHIE team who worked on this recent publication featured by @MSKLibrary: A Bayesian active learning platform for scalable combination drug screens dx.doi.org/10.1038/s414...
Reposted by Mark Polk
Happy new year!
2024 was a productive year, and 2025 stands to be even more exciting!
Our thanks to everyone who helped, and some reflections: foldingathome.org/2025/01/09/h...
2024 was a productive year, and 2025 stands to be even more exciting!
Our thanks to everyone who helped, and some reflections: foldingathome.org/2025/01/09/h...
Happy new year 2025! – Folding@home
foldingathome.org
January 10, 2025 at 2:01 PM
Happy new year!
2024 was a productive year, and 2025 stands to be even more exciting!
Our thanks to everyone who helped, and some reflections: foldingathome.org/2025/01/09/h...
2024 was a productive year, and 2025 stands to be even more exciting!
Our thanks to everyone who helped, and some reflections: foldingathome.org/2025/01/09/h...
So grateful to have had the opportunity to chat science in Berlin with our collaborators in the Volkamer Lab and in the UK at the Oxford Centre for Medicines Discovery and the Diamond Light Source. Many thanks to DLS for the tour!

December 10, 2024 at 5:12 PM
So grateful to have had the opportunity to chat science in Berlin with our collaborators in the Volkamer Lab and in the UK at the Oxford Centre for Medicines Discovery and the Diamond Light Source. Many thanks to DLS for the tour!
Reposted by Mark Polk
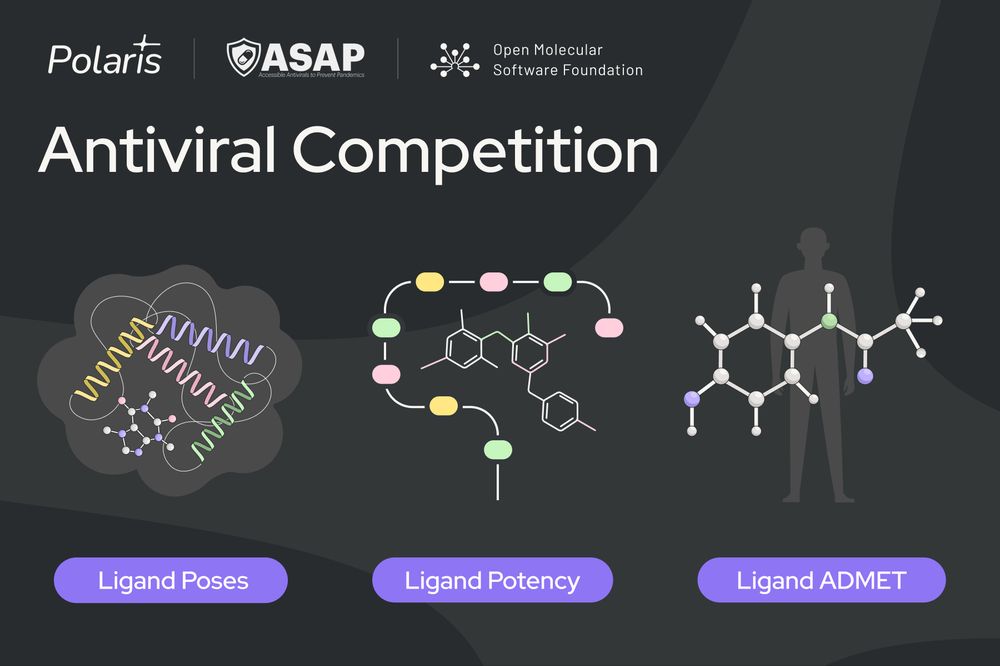
🦠 We’re excited to announce our first competition in partnership with @asapdiscovery.bsky.social and @omsf.io!
Test your skills across three sub-challenges revolving around SARS-CoV-2 and MERS-CoV Mpro🧵
Full details: polarishub.io/competitions
Blog: polarishub.io/blog/antivir...
Test your skills across three sub-challenges revolving around SARS-CoV-2 and MERS-CoV Mpro🧵
Full details: polarishub.io/competitions
Blog: polarishub.io/blog/antivir...
December 3, 2024 at 3:13 PM
🦠 We’re excited to announce our first competition in partnership with @asapdiscovery.bsky.social and @omsf.io!
Test your skills across three sub-challenges revolving around SARS-CoV-2 and MERS-CoV Mpro🧵
Full details: polarishub.io/competitions
Blog: polarishub.io/blog/antivir...
Test your skills across three sub-challenges revolving around SARS-CoV-2 and MERS-CoV Mpro🧵
Full details: polarishub.io/competitions
Blog: polarishub.io/blog/antivir...
Automated plate handling in the Chodera Lab, part of our effort to study kinase inhibitors in high throughput.
November 27, 2024 at 4:44 PM
Automated plate handling in the Chodera Lab, part of our effort to study kinase inhibitors in high throughput.

