
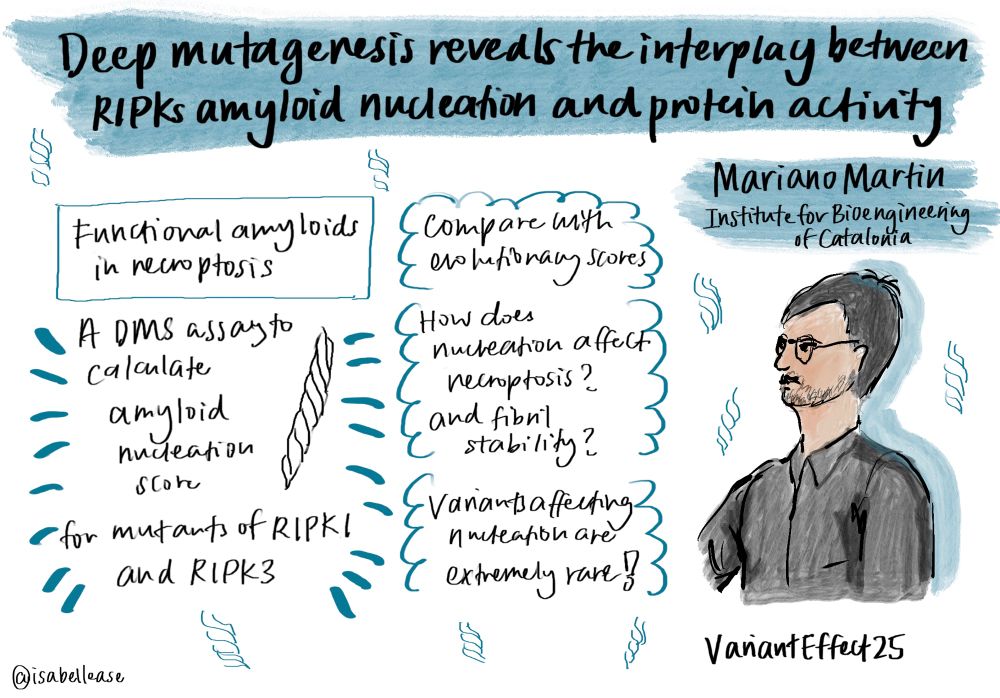
Deep mutagenesis reveals the interplay between RIPKs amyloid nucleation and protein activity

Deep mutagenesis reveals the interplay between RIPKs amyloid nucleation and protein activity








Find out more this week on @pnas.org!
www.pnas.org/doi/10.1073/...



Find out more this week on @pnas.org!
www.pnas.org/doi/10.1073/...
We studied MCT8 deficiency via genomic, functional & deep learning integration.
🔗 www.nature.com/articles/s41...

We studied MCT8 deficiency via genomic, functional & deep learning integration.
🔗 www.nature.com/articles/s41...
We quantified ~100k sequences to train CANYA, an interpretable deep learning model for predicting amyloid formation.
🔗 www.science.org/doi/10.1126/...

We quantified ~100k sequences to train CANYA, an interpretable deep learning model for predicting amyloid formation.
🔗 www.science.org/doi/10.1126/...
We used deep mutational scanning & random extensions to uncover how Bri2 becomes amyloidogenic upon C-terminal extension.
🔗 www.pnas.org/doi/10.1073/...
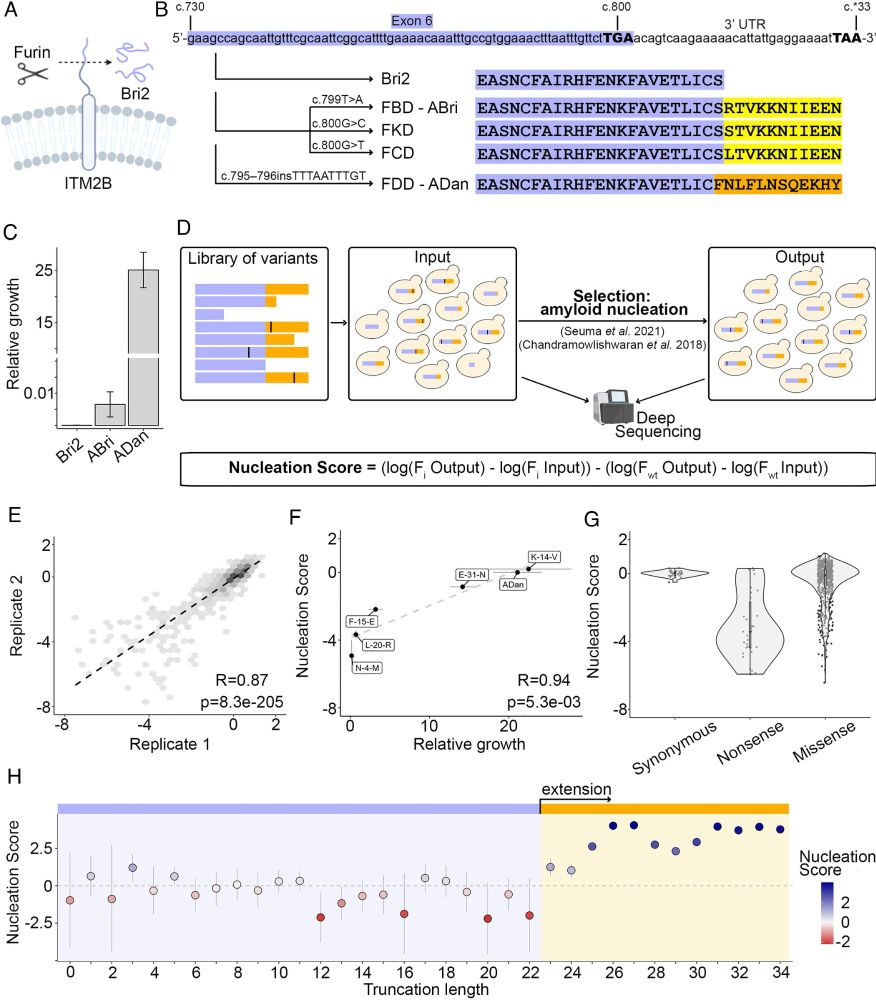
We used deep mutational scanning & random extensions to uncover how Bri2 becomes amyloidogenic upon C-terminal extension.
🔗 www.pnas.org/doi/10.1073/...
www.varianteffect.org/seminar-series

www.varianteffect.org/seminar-series
🌍 @ibecbarcelona.eu @crg.eu @varianteffect.bsky.social
events.ibecbarcelona.eu/mutational-s...

🌍 @ibecbarcelona.eu @crg.eu @varianteffect.bsky.social
events.ibecbarcelona.eu/mutational-s...

