
www.biorxiv.org/content/10.1...
(1/n)

www.biorxiv.org/content/10.1...
(1/n)
The E2G Portal! e2g.stanford.edu
This collates our predictions of enhancer-gene regulatory interactions across >1,600 cell types and tissues.
Uses cases 👇
1/
The E2G Portal! e2g.stanford.edu
This collates our predictions of enhancer-gene regulatory interactions across >1,600 cell types and tissues.
Uses cases 👇
1/
◼️ They do not offer compelling gains over baseline models
Their performance is inconsistent and requires much more compute.
arxiv.org/abs/2412.05430

◼️ They do not offer compelling gains over baseline models
Their performance is inconsistent and requires much more compute.
arxiv.org/abs/2412.05430
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
@lauradmartens.bsky.social @johahi.bsky.social @kipoizoo.bsky.social
Last preprint version:
www.biorxiv.org/content/10.1...
@lauradmartens.bsky.social @johahi.bsky.social @kipoizoo.bsky.social
Last preprint version:
www.biorxiv.org/content/10.1...


The ENCODE 4 expanded registry of regulatory elements
- 2.35M 🧍 human cCREs
- 927k 🐭 mouse cCREs
www.biorxiv.org/content/10.1...
Led by @moorejille.bsky.social, this preprint summarizes data and analyses generated by hundreds of contributors across ENCODE 4
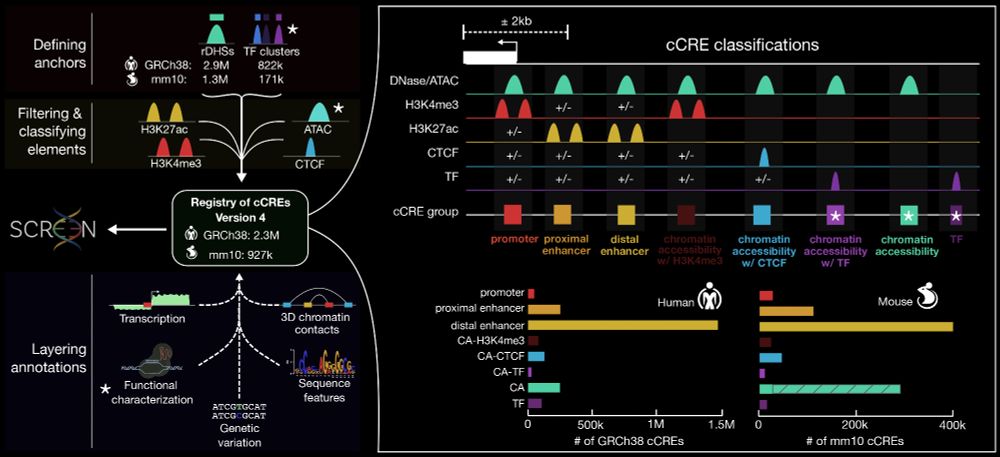
The ENCODE 4 expanded registry of regulatory elements
- 2.35M 🧍 human cCREs
- 927k 🐭 mouse cCREs
www.biorxiv.org/content/10.1...
Led by @moorejille.bsky.social, this preprint summarizes data and analyses generated by hundreds of contributors across ENCODE 4
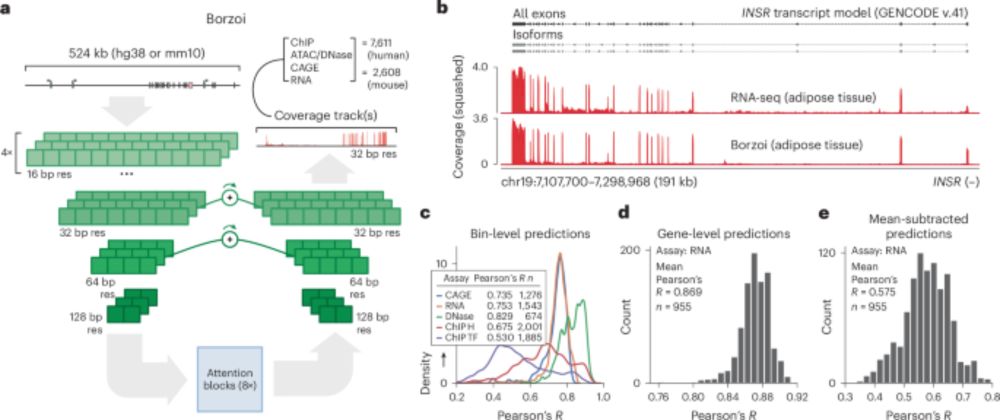
www.biorxiv.org/content/10.1...
Huge congrats to Anusri! This was quite a slog (for both of us) but we r very proud of this one! It is a long read but worth it IMHO. Methods r in the supp. materials. Bluetorial coming soon below 1/
www.biorxiv.org/content/10.1...
Huge congrats to Anusri! This was quite a slog (for both of us) but we r very proud of this one! It is a long read but worth it IMHO. Methods r in the supp. materials. Bluetorial coming soon below 1/
We usually find that most fine-mapped variants do not fall within coding or regulatory regions. Is it a limitation of epigenomics or a limitation of fine-mapping? Please share your thoughts!
We usually find that most fine-mapped variants do not fall within coding or regulatory regions. Is it a limitation of epigenomics or a limitation of fine-mapping? Please share your thoughts!
github.com/kristen-schn...

github.com/kristen-schn...

This list is pretty dang awesome: github.com/KenneyNL/Ado...
#gamedev

This list is pretty dang awesome: github.com/KenneyNL/Ado...
#gamedev
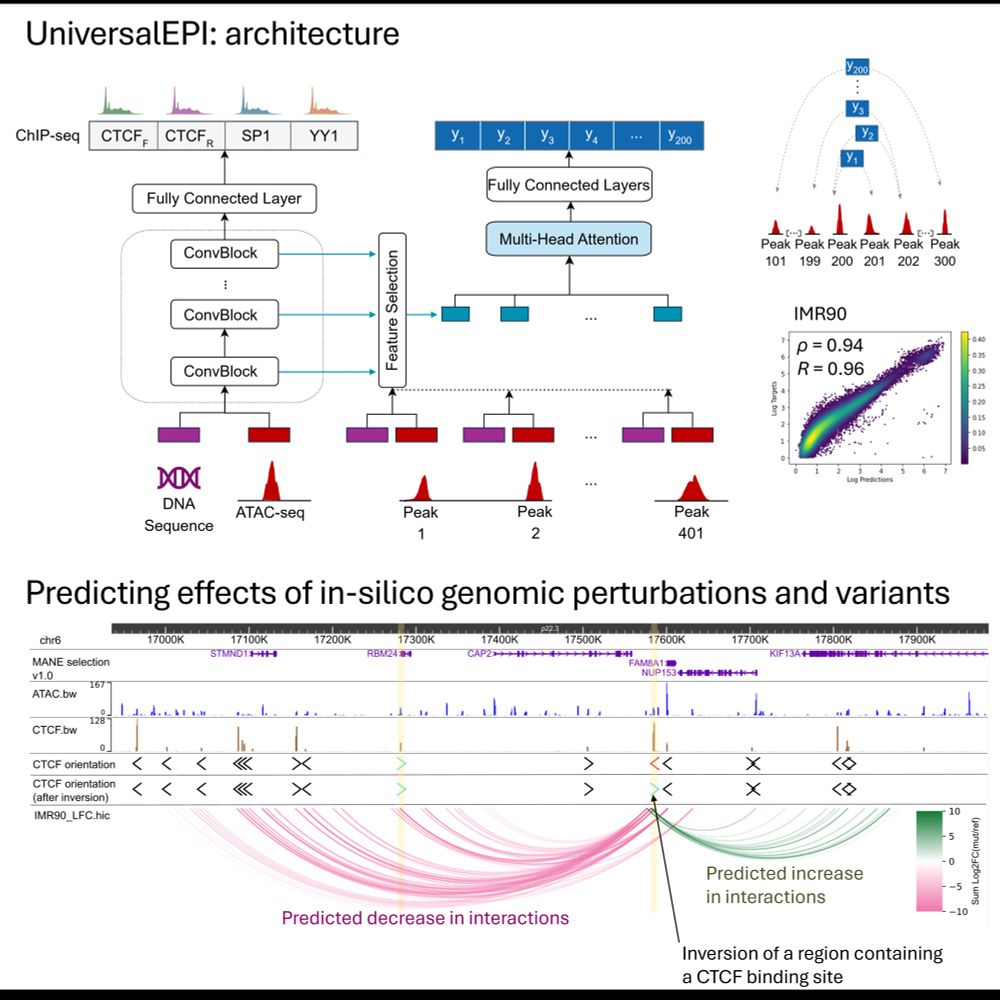
arxiv.org/abs/2411.04118

www.nature.com/articles/s41...
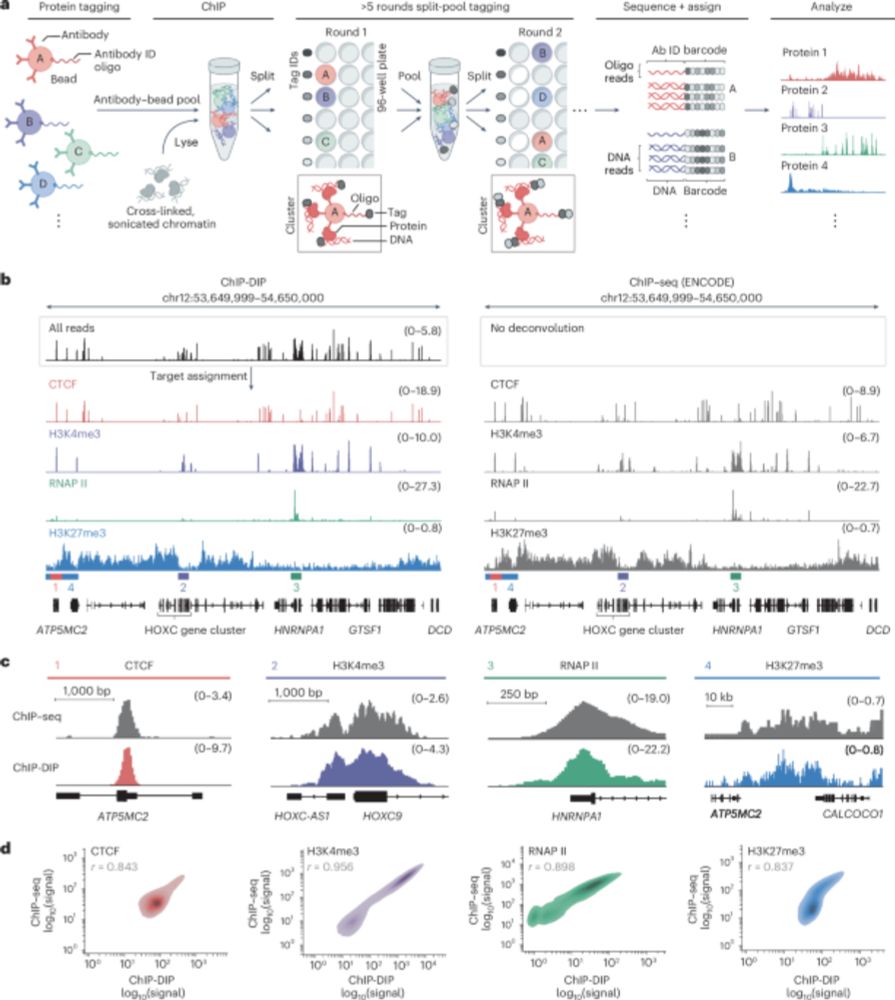
www.nature.com/articles/s41...


