

🔗 rdcu.be/eiHRG
@sgn.one @leibnizlib.bsky.social #biodiversity #genomics

🔗 rdcu.be/eiHRG
@sgn.one @leibnizlib.bsky.social #biodiversity #genomics




academic.oup.com/bioinformati...

academic.oup.com/bioinformati...
pixy.readthedocs.io
www.popgen.dk/angsd/index....
github.com/RILAB/mop

pixy.readthedocs.io
www.popgen.dk/angsd/index....
github.com/RILAB/mop
press.princeton.edu/ideas/what-i...
#Science #Nature #bees #AnimalBehavior


press.princeton.edu/ideas/what-i...
#Science #Nature #bees #AnimalBehavior



THE INFINITE EXTENT. A book about life at different scales--of size, time, geography, longevity, connectivity, individuality, & more.
I see it as the conclusion of a trilogy of books.
More details: buttondown.email/edyong209/ar...
THE INFINITE EXTENT. A book about life at different scales--of size, time, geography, longevity, connectivity, individuality, & more.
I see it as the conclusion of a trilogy of books.
More details: buttondown.email/edyong209/ar...


academic.oup.com/sysbio/advan...
🧪🦀🦑#evosky
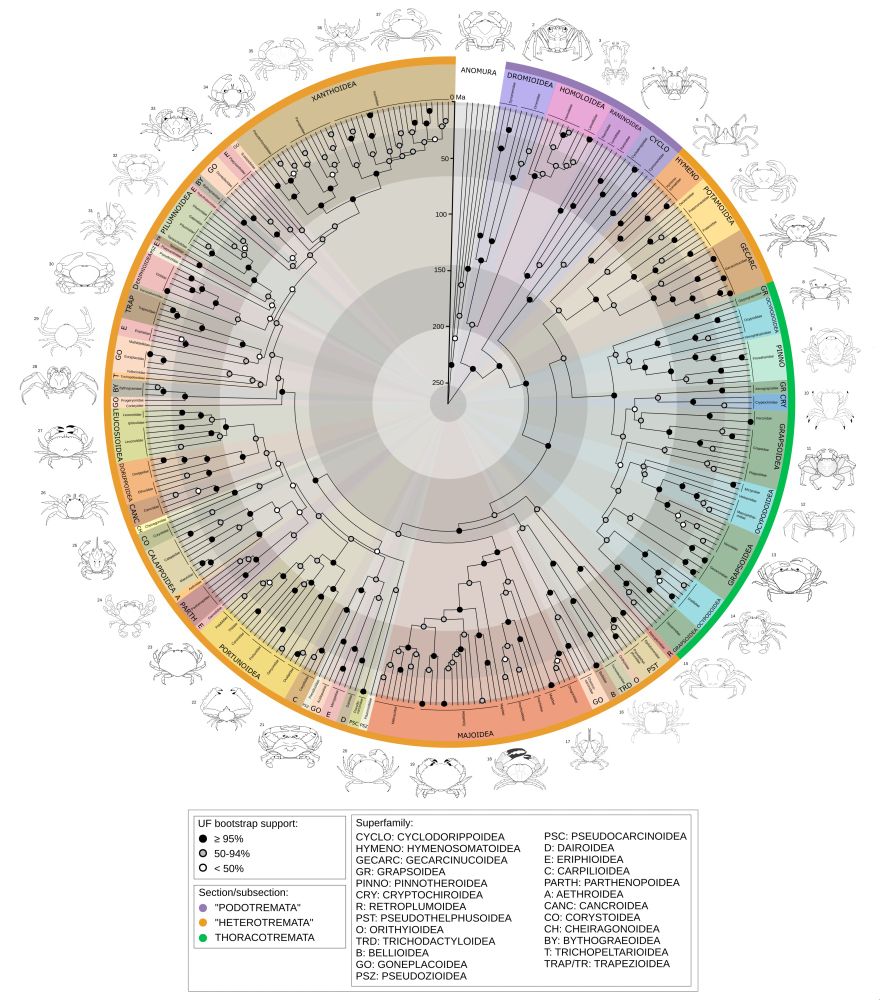
academic.oup.com/sysbio/advan...
🧪🦀🦑#evosky


