Spatial omics, neurodevelopment and more in Bayraktar lab, @sangerinstitute.bsky.social
With amazing co-authors Alexander & Fani we are passionate about neurodevelopmental genetics and this work really showed us the power of uncovering new genetics with spatial transcriptomics!
🧵👇 Background / motivation / key results

With amazing co-authors Alexander & Fani we are passionate about neurodevelopmental genetics and this work really showed us the power of uncovering new genetics with spatial transcriptomics!
🧵👇 Background / motivation / key results
Join us to generate & analyse wealths of Xenium data:
- Tech specialist: sanger.wd103.myworkdayjobs.com/en-US/Wellco...
- AI postdoc: sanger.wd103.myworkdayjobs.com/en-US/Wellco...
Join us to generate & analyse wealths of Xenium data:
- Tech specialist: sanger.wd103.myworkdayjobs.com/en-US/Wellco...
- AI postdoc: sanger.wd103.myworkdayjobs.com/en-US/Wellco...
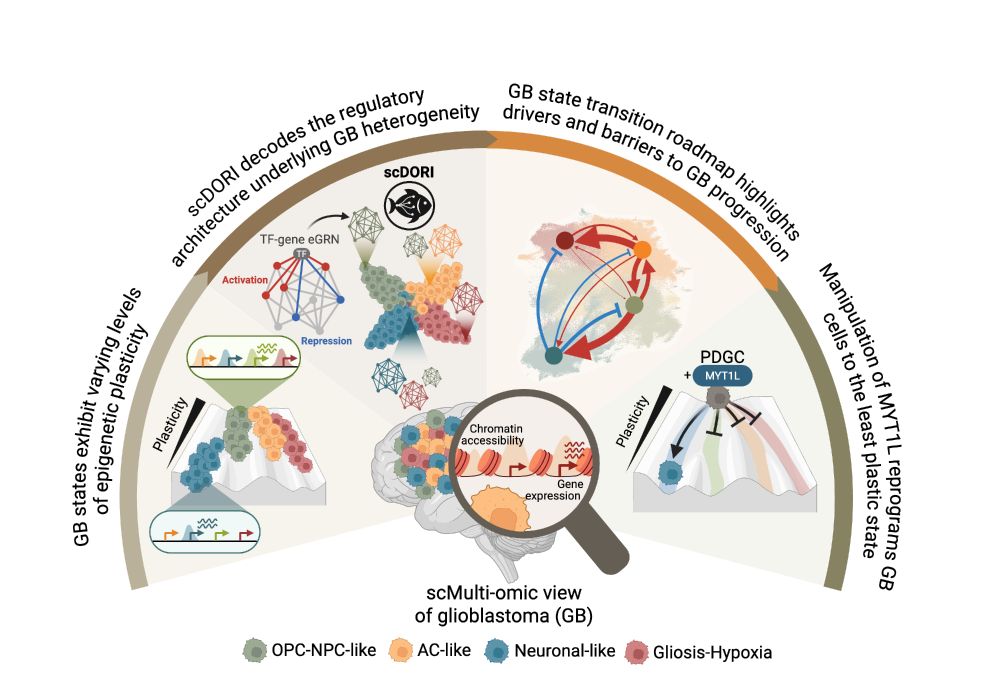
We infer metabolic activity from single-cell & spatial transcriptomics 🧬
Read the pre-print here👇
📄 doi.org/10.1101/2025...
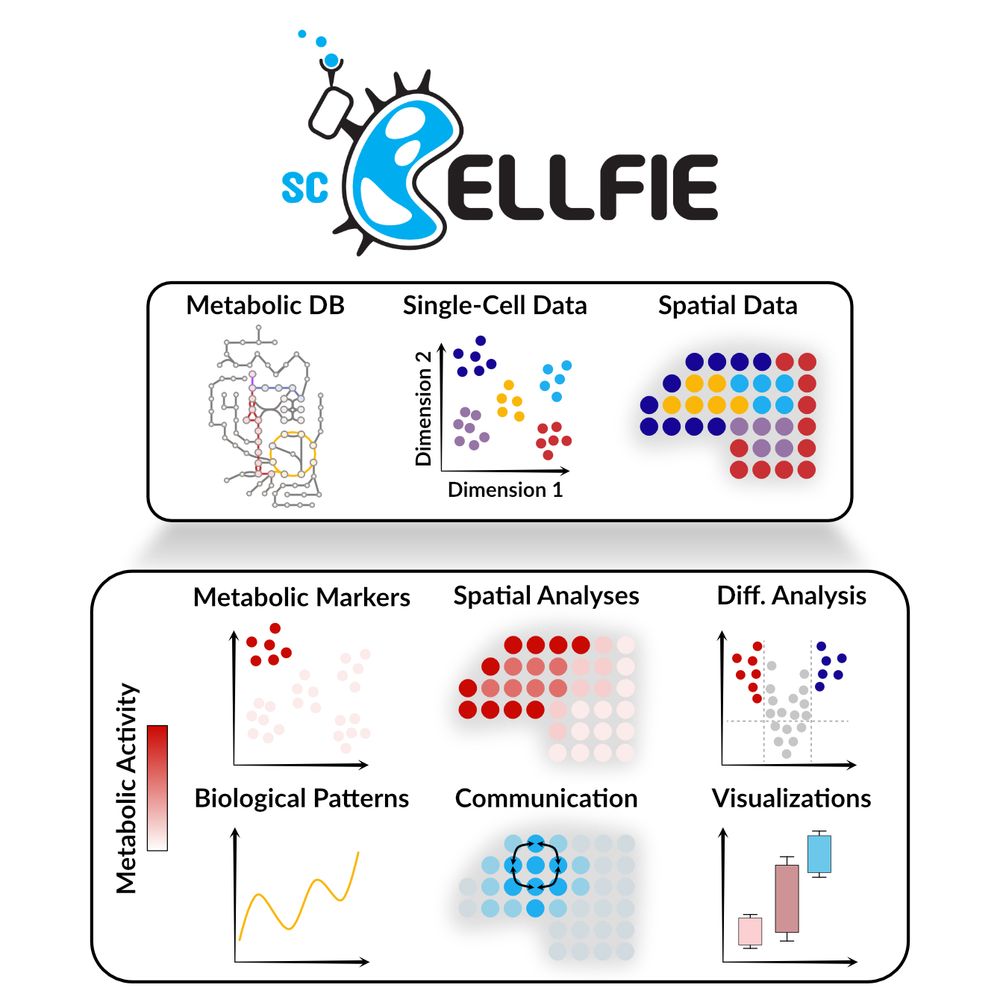
We infer metabolic activity from single-cell & spatial transcriptomics 🧬
Read the pre-print here👇
📄 doi.org/10.1101/2025...
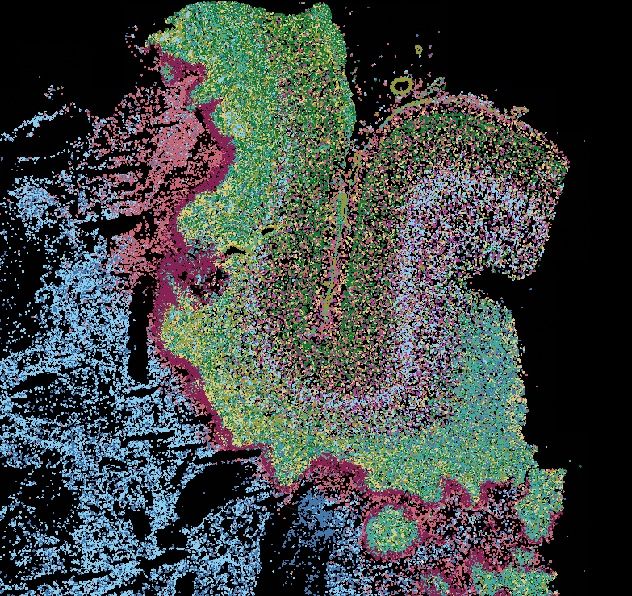
Using single-cell & histopathology-informed #SpatialTranscriptomics, we uncovered key biomarkers & immune patterns. Thanks to collaborators, UKCIC & @humancellatlas.org
By understanding how our lungs respond to the virus over time, this new knowledge may help identify future therapies for COVID-19.
www.sanger.ac.uk/news_item/ce...

Using single-cell & histopathology-informed #SpatialTranscriptomics, we uncovered key biomarkers & immune patterns. Thanks to collaborators, UKCIC & @humancellatlas.org
Alexander Aivazidis has been a great colleague in our lab and it's amazing to see the capstone of his PhD is now published.
here: bit.ly/3Diarf5

Alexander Aivazidis has been a great colleague in our lab and it's amazing to see the capstone of his PhD is now published.
Two amazing people are running for charities, please support them!
Two amazing people are running for charities, please support them!
2/8

