www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

www.biorxiv.org/cgi/content/...
www.biorxiv.org/cgi/content/...
BRD4-NUT forms liquid-like condensates that locally constrain nucleosomes via BRD4-mediated crosslinking— physical control of #chromatin by #LLPS transcription condensates. @semeigazin.bsky.social @katsuminami.bsky.social @masaashimazoe.bsky.social
BRD4-NUT forms liquid-like condensates that locally constrain nucleosomes via BRD4-mediated crosslinking— physical control of #chromatin by #LLPS transcription condensates. @semeigazin.bsky.social @katsuminami.bsky.social @masaashimazoe.bsky.social
@katsuminami.bsky.social @semeigazin.bsky.social @kakonakazato.bsky.social discuss DNA accessibility in euchromatin and heterochromatin of live cells.
Free access: authors.elsevier.com/a/1lHgb54HFZ...
Also check our previous work: www.science.org/doi/10.1126/...
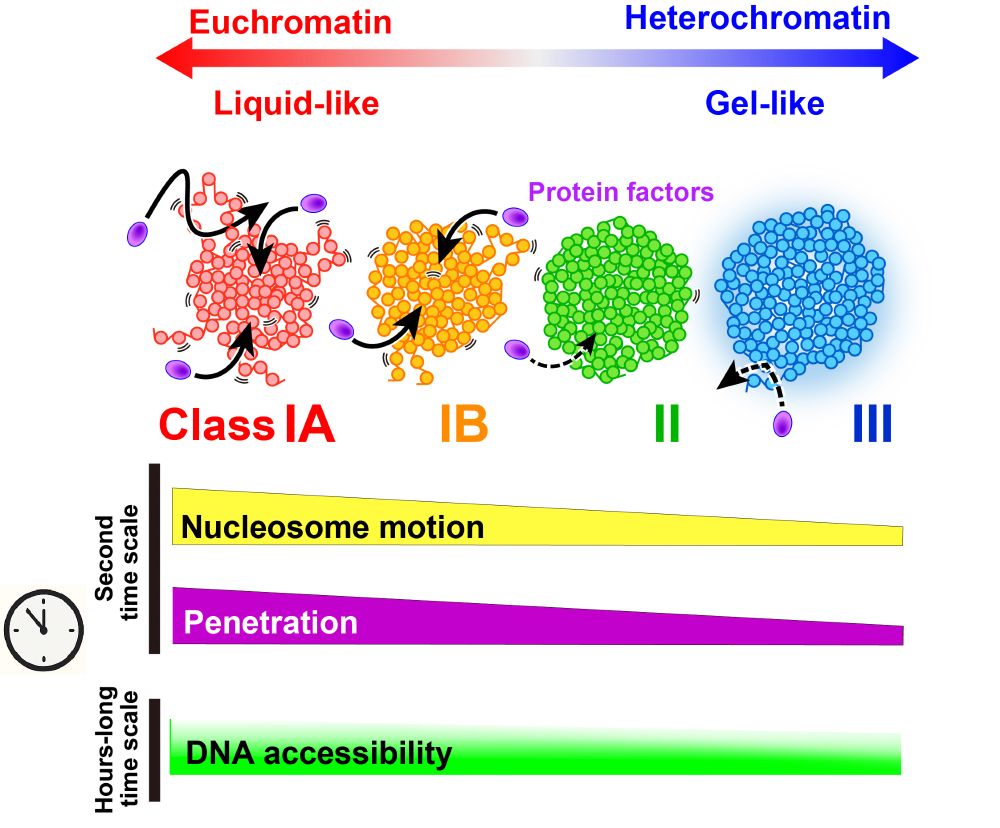
@katsuminami.bsky.social @semeigazin.bsky.social @kakonakazato.bsky.social discuss DNA accessibility in euchromatin and heterochromatin of live cells.
Free access: authors.elsevier.com/a/1lHgb54HFZ...
Also check our previous work: www.science.org/doi/10.1126/...
www.nig.ac.jp/nig/career-d...
🧬We’re seeking motivated postdocs to join our lab. 🔬We study chromatin behavior in living cells using advanced live-cell imaging and quantitative analysis:
www.science.org/doi/10.1126/...
doi.org/10.2183/pjab...
Interested? Please apply!

www.nig.ac.jp/nig/career-d...
🧬We’re seeking motivated postdocs to join our lab. 🔬We study chromatin behavior in living cells using advanced live-cell imaging and quantitative analysis:
www.science.org/doi/10.1126/...
doi.org/10.2183/pjab...
Interested? Please apply!
🔬 Original paper: www.science.org/doi/full/10....

🔬 Original paper: www.science.org/doi/full/10....
doi.org/10.2183/pjab...
The textbook view of #chromatin as 30-nm fibers is fading.
A new paradigm: chromatin forms #liquid-like domains—locally dynamic, yet globally stable at the chromosome level (viscoelastic)—supporting genome function. ✨
doi.org/10.2183/pjab...
The textbook view of #chromatin as 30-nm fibers is fading.
A new paradigm: chromatin forms #liquid-like domains—locally dynamic, yet globally stable at the chromosome level (viscoelastic)—supporting genome function. ✨
Our Repli-Histo labeling revealed the distinct physical properties of euchromatin (liquid-like) and heterochromatin (gel-like) and their implications for replication timing.
www.science.org/doi/10.1126/...
🧬Our Repli-Histo labeling marks nucleosomes in euchromatin and heterochromatin in live human cells.
🔍 @katsuminami.bsky.social et al. have developed a chromatin behavior atlas within the nucleus. 1/2
Our Repli-Histo labeling revealed the distinct physical properties of euchromatin (liquid-like) and heterochromatin (gel-like) and their implications for replication timing.
🧪 More euchromatic regions (i.e., earlier-replicated regions) show greater nucleosome motion.
🎉 Congrats @katsuminami.bsky.social @kakonakazato.bsky.social and all collaborators!
2/2
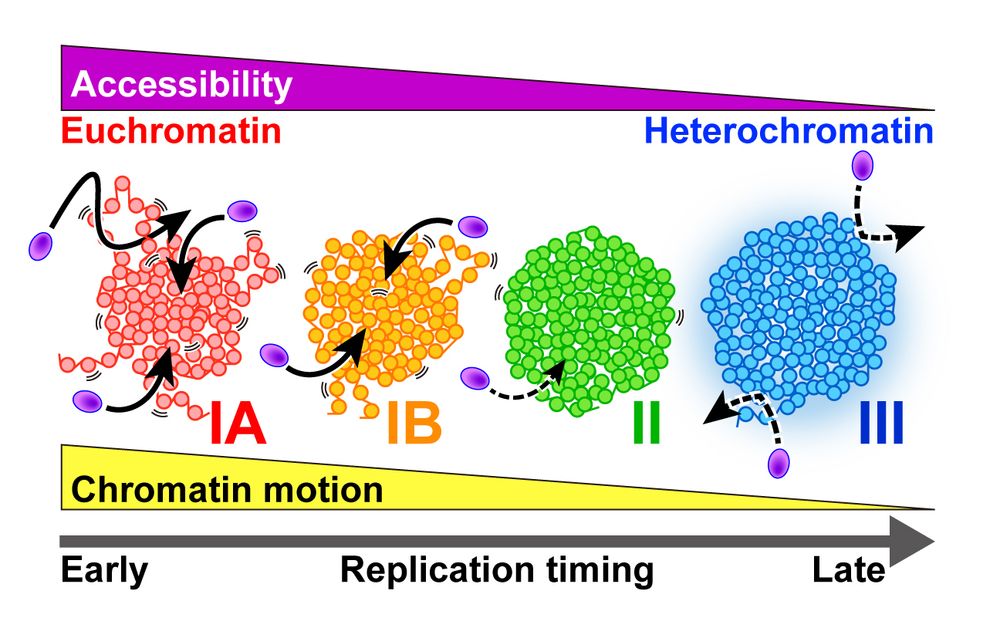
🧪 More euchromatic regions (i.e., earlier-replicated regions) show greater nucleosome motion.
🎉 Congrats @katsuminami.bsky.social @kakonakazato.bsky.social and all collaborators!
2/2
www.science.org/doi/10.1126/...
🧬Our Repli-Histo labeling marks nucleosomes in euchromatin and heterochromatin in live human cells.
🔍 @katsuminami.bsky.social et al. have developed a chromatin behavior atlas within the nucleus. 1/2
www.science.org/doi/10.1126/...
🧬Our Repli-Histo labeling marks nucleosomes in euchromatin and heterochromatin in live human cells.
🔍 @katsuminami.bsky.social et al. have developed a chromatin behavior atlas within the nucleus. 1/2
@masaashimazoe.bsky.social et al. reveal that linker histone H1 acts as a liquid-like glue to organize chromatin in living cells. 🎉 Fantastic collab with @rcollepardo.bsky.social @janhuemar.bsky.social and others—huge thanks! 🙌 1/
@masaashimazoe.bsky.social et al. reveal that linker histone H1 acts as a liquid-like glue to organize chromatin in living cells. 🎉 Fantastic collab with @rcollepardo.bsky.social @janhuemar.bsky.social and others—huge thanks! 🙌 1/
We critically discuss the domain formation mechanism from a physical perspective, including #phase-separation and #condensation.
📥 Free-download link:
authors.elsevier.com/a/1keGn,LqAr...
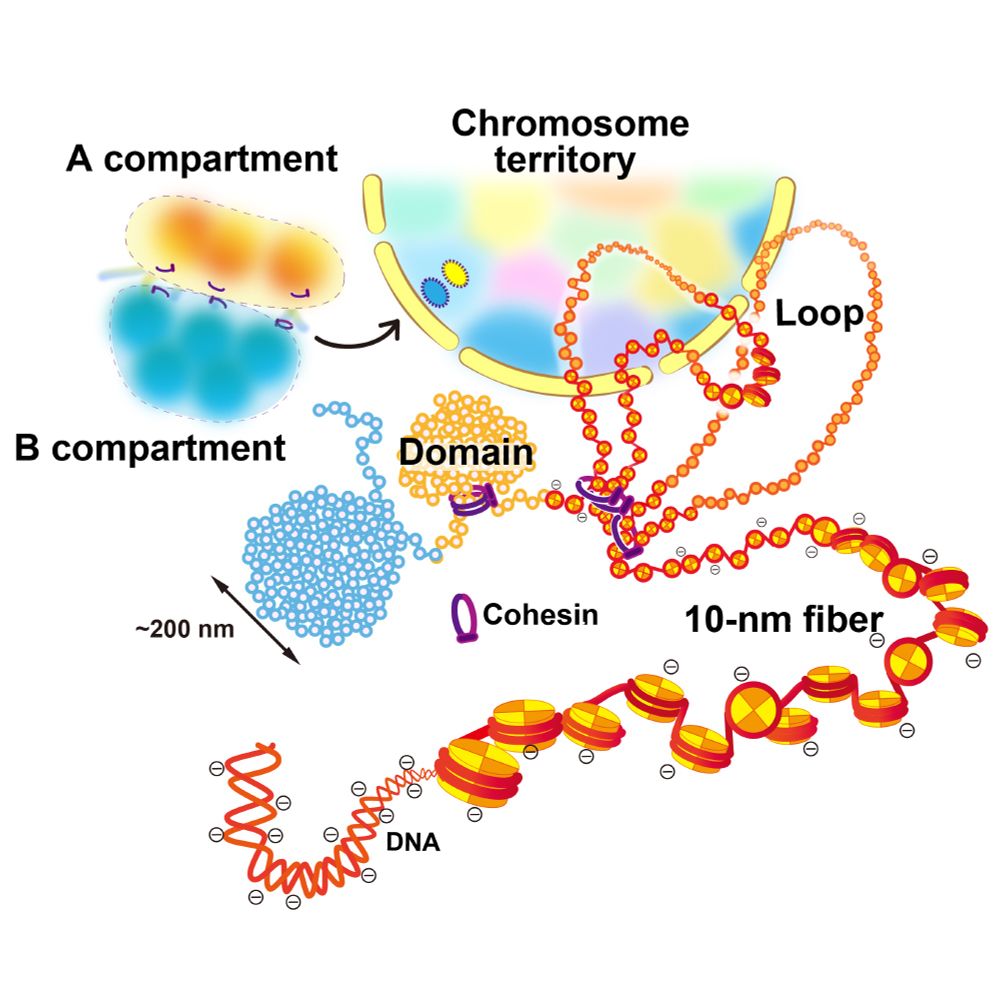
We critically discuss the domain formation mechanism from a physical perspective, including #phase-separation and #condensation.
📥 Free-download link:
authors.elsevier.com/a/1keGn,LqAr...
https://go.nature.com/4k8AdCX

https://go.nature.com/4k8AdCX
(X: katsu_s_minami)
(X: katsu_s_minami)

