* Download: zenodo.org/records/1490...
* More info: github.com/lh3/panmask

* Download: zenodo.org/records/1490...
* More info: github.com/lh3/panmask


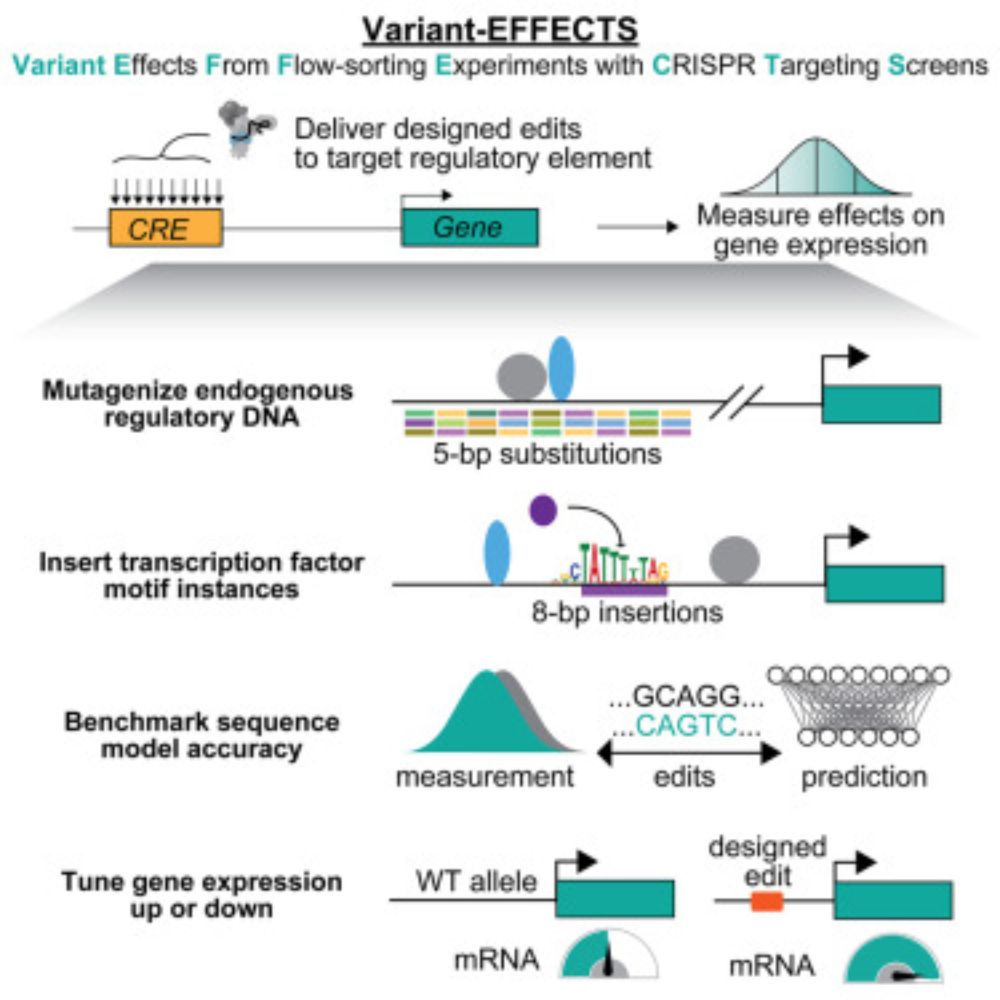
API: github.com/google-deepm...
Docs: www.alphagenomedocs.com
Community: www.alphagenomecommunity.com
API: github.com/google-deepm...
Docs: www.alphagenomedocs.com
Community: www.alphagenomecommunity.com



www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Free to read here: rdcu.be/dQEND

Free to read here: rdcu.be/dQEND
Human vs macaque:
▶️2133 gained (2.7%), 2418 lost (3.1%) CTCF loops in B-cells
▶️5873 gained (7.3%), 6708 lost (8.3%) in neurons
▶️implications for ASD and immunity

Human vs macaque:
▶️2133 gained (2.7%), 2418 lost (3.1%) CTCF loops in B-cells
▶️5873 gained (7.3%), 6708 lost (8.3%) in neurons
▶️implications for ASD and immunity
Join us to learn genome assemby&annotation, variant detection, and evolutionary analysis with lots of hands-on sessions! @nanoporetech.com @pacbio.bsky.social @sedlazeck.bsky.social
www.physalia-courses.org/courses-work...

Join us to learn genome assemby&annotation, variant detection, and evolutionary analysis with lots of hands-on sessions! @nanoporetech.com @pacbio.bsky.social @sedlazeck.bsky.social
www.physalia-courses.org/courses-work...
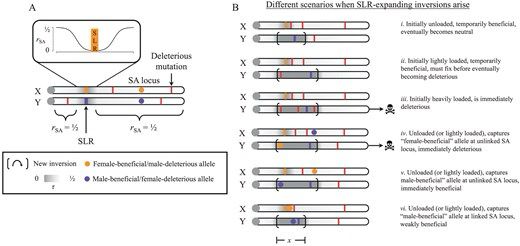
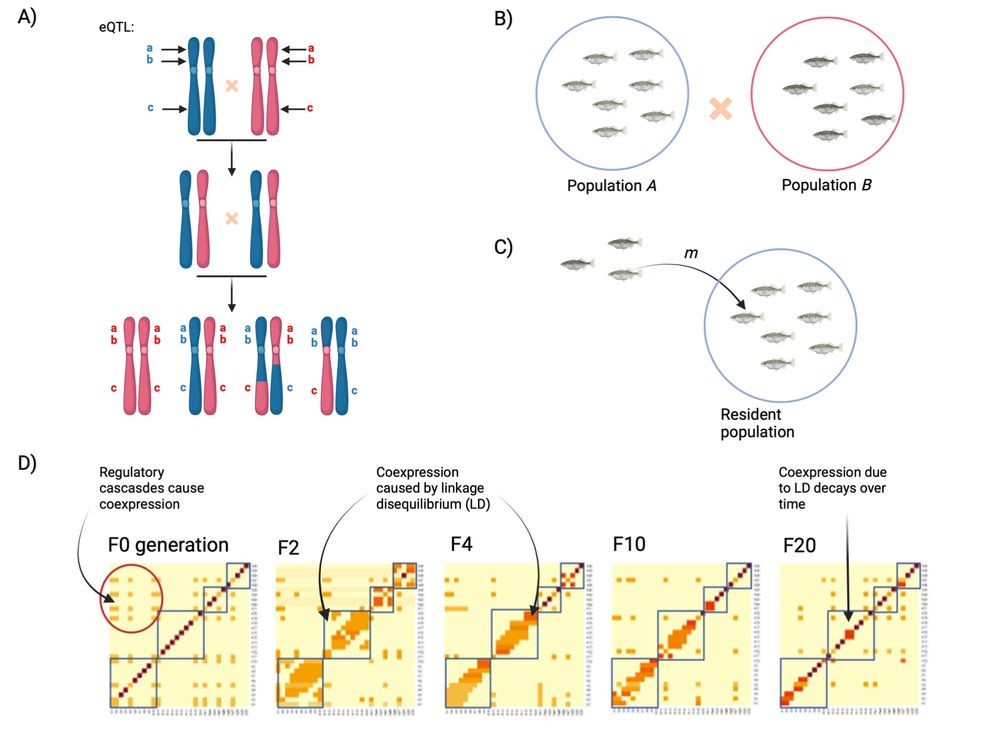
academic.oup.com/genetics/adv...
We ask what gene flow & introgression do to gene coexpression networks. The answer is: a lot.
academic.oup.com/genetics/adv...
We ask what gene flow & introgression do to gene coexpression networks. The answer is: a lot.
key point: 2/3rds of TF isos differ in properties like DNA binding & transcriptional activity
many are "negative regulators" & misexpressed in cancer
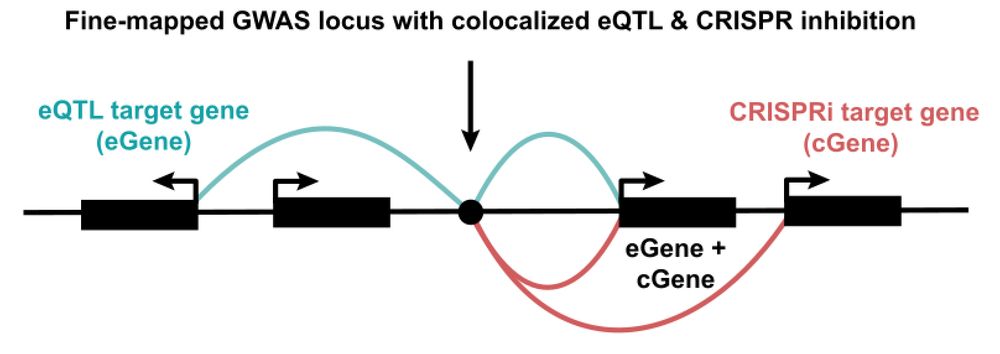
www.sciencedirect.com/science/arti...
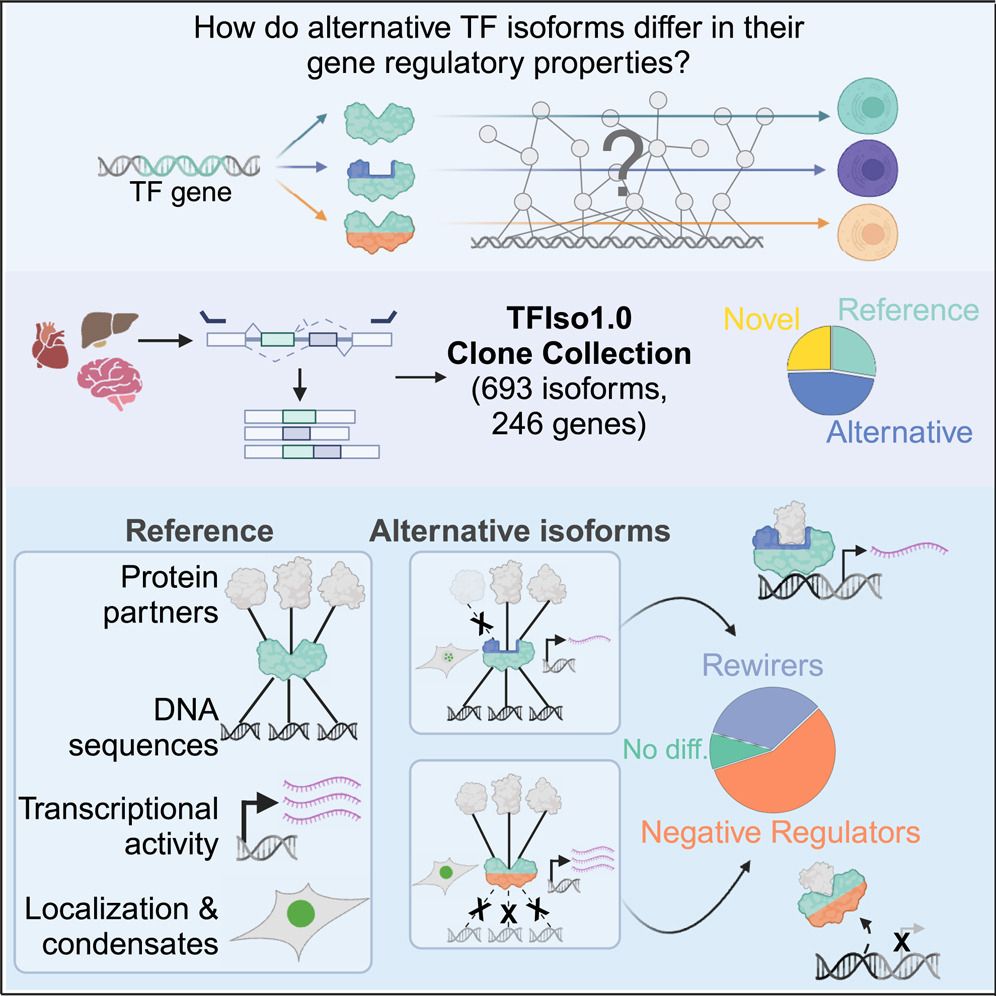
key point: 2/3rds of TF isos differ in properties like DNA binding & transcriptional activity
many are "negative regulators" & misexpressed in cancer
www.sciencedirect.com/science/arti...
Check out the database resource, as STRchive "streamlines TR variant interpretation at disease-associated loci."
strchive.org
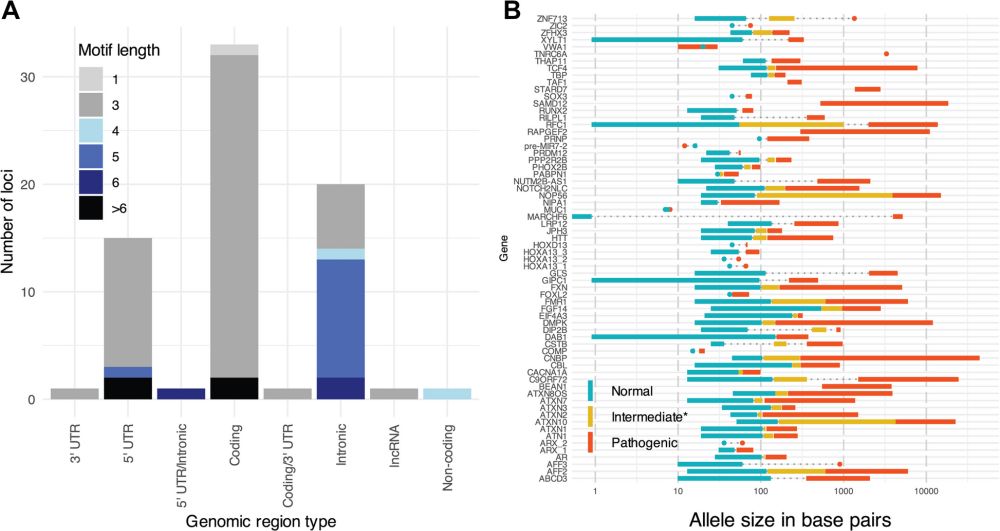
Check out the database resource, as STRchive "streamlines TR variant interpretation at disease-associated loci."
strchive.org




We investigate how well you can call variants directly from genome assemblies compared to traditional read-based variant calling.
Read it here: www.biorxiv.org/content/10.1...
Data & code: github.com/rrwick/Are-r...
(1/8)

We investigate how well you can call variants directly from genome assemblies compared to traditional read-based variant calling.
Read it here: www.biorxiv.org/content/10.1...
Data & code: github.com/rrwick/Are-r...
(1/8)




www.science.org/doi/10.1126/...
www.science.org/doi/10.1126/...
www.science.org/doi/10.1126/...
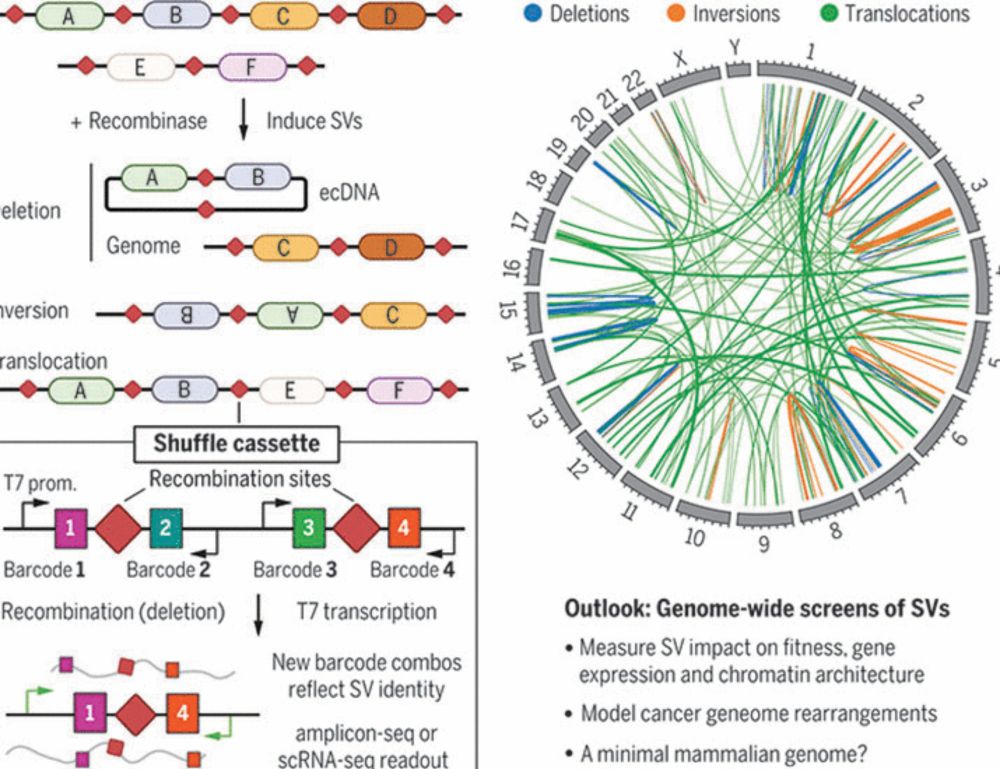
www.science.org/doi/10.1126/...