
Jorge Sastre Domínguez
@jorgesastred.bsky.social
PhD student in the Plasmid Biology and Evolution (PBE) and Evolution of Microbes and Mobile Genetic Elements labs.
Bioinformatics 💻 Evolutionary Biology 🦠 Antimicrobial resistance 💊
📍CNB - CSIC
Bioinformatics 💻 Evolutionary Biology 🦠 Antimicrobial resistance 💊
📍CNB - CSIC
Reposted by Jorge Sastre Domínguez
🚨 Excited to share our new paper is out! 🎉
We show how interactions within gut microbiomes allow certain antibiotic-resistant E. coli strains to persist even without antibiotics, helping explain how resistance is maintained in the human gut.
Now published in @natcomms.nature.com rdcu.be/eOf63
We show how interactions within gut microbiomes allow certain antibiotic-resistant E. coli strains to persist even without antibiotics, helping explain how resistance is maintained in the human gut.
Now published in @natcomms.nature.com rdcu.be/eOf63

Multi-layered ecological interactions determine growth of clinical antibiotic-resistant strains within human microbiomes
Nature Communications - The role of ecological factors in modulating the spread of antibiotic-resistance bacteria in the gut remains unclear. Here, the authors use anaerobic microcosms to study the...
rdcu.be
November 7, 2025 at 9:15 AM
🚨 Excited to share our new paper is out! 🎉
We show how interactions within gut microbiomes allow certain antibiotic-resistant E. coli strains to persist even without antibiotics, helping explain how resistance is maintained in the human gut.
Now published in @natcomms.nature.com rdcu.be/eOf63
We show how interactions within gut microbiomes allow certain antibiotic-resistant E. coli strains to persist even without antibiotics, helping explain how resistance is maintained in the human gut.
Now published in @natcomms.nature.com rdcu.be/eOf63
Reposted by Jorge Sastre Domínguez
So happy to share this! Bacteriocins were first discovered over 100 years ago, but what do they actually do? We look at >1000 bacteriocin plasmids and find links to virulence and antimicrobial resistance, and frequent bacteriocin sharing in Enterobacteriaceae.
www.nature.com/articles/s41...
www.nature.com/articles/s41...

Bacterial warfare is associated with virulence and antimicrobial resistance - Nature Communications
Bacteria employ a range of competition systems that deliver toxins to inhibit competing strains. This study shows that these systems are particularly important for the ecology of virulent and antibiot...
www.nature.com
November 5, 2025 at 7:32 AM
So happy to share this! Bacteriocins were first discovered over 100 years ago, but what do they actually do? We look at >1000 bacteriocin plasmids and find links to virulence and antimicrobial resistance, and frequent bacteriocin sharing in Enterobacteriaceae.
www.nature.com/articles/s41...
www.nature.com/articles/s41...
Reposted by Jorge Sastre Domínguez
Reposted by Jorge Sastre Domínguez
@prczhaoyansong.bsky.social’s deep dive into the dark matter of compost communities is now out 🎉 Genomic islands hijack jumbo phages—whose capsids enable transfer of large tracts of DNA—shedding new light on the scale & scope of phage-mediated gene flow 😎
www.pnas.org/doi/10.1073/...
www.pnas.org/doi/10.1073/...

Jumbo phage–mediated transduction of genomic islands | PNAS
Bacteria acquire new genes by horizontal gene transfer, typically mediated by mobile
genetic elements (MGEs). While plasmids, bacteriophages, and c...
www.pnas.org
October 28, 2025 at 6:36 PM
@prczhaoyansong.bsky.social’s deep dive into the dark matter of compost communities is now out 🎉 Genomic islands hijack jumbo phages—whose capsids enable transfer of large tracts of DNA—shedding new light on the scale & scope of phage-mediated gene flow 😎
www.pnas.org/doi/10.1073/...
www.pnas.org/doi/10.1073/...
Reposted by Jorge Sastre Domínguez
Do plasmids really move around that much? Well, maybe not always
Thrilled to have contributed to this story with two of my favourite microbiologists: @jrpenades.bsky.social & @sanmillan.bsky.social
This great work was led by Akshay Sabnis & @wfigueroac3.bsky.social
www.cell.com/cell-reports...
Thrilled to have contributed to this story with two of my favourite microbiologists: @jrpenades.bsky.social & @sanmillan.bsky.social
This great work was led by Akshay Sabnis & @wfigueroac3.bsky.social
www.cell.com/cell-reports...

Non-conjugative plasmids limit their mobility to persist in nature
Sabnis et al. explain why non-conjugative plasmids move at a low rate in nature. While
increased mobility can easily evolve by incorporating phage DNA into plasmids, this
is disadvantageous because it...
www.cell.com
October 22, 2025 at 5:47 PM
Do plasmids really move around that much? Well, maybe not always
Thrilled to have contributed to this story with two of my favourite microbiologists: @jrpenades.bsky.social & @sanmillan.bsky.social
This great work was led by Akshay Sabnis & @wfigueroac3.bsky.social
www.cell.com/cell-reports...
Thrilled to have contributed to this story with two of my favourite microbiologists: @jrpenades.bsky.social & @sanmillan.bsky.social
This great work was led by Akshay Sabnis & @wfigueroac3.bsky.social
www.cell.com/cell-reports...
Reposted by Jorge Sastre Domínguez
New paper with my (amazing) friend and mentor @jrpenades.bsky.social
Really looking forward to see what plasmid aficionados think of this one!!
With @asantoslopez.bsky.social @wfigueroac3.bsky.social Akshay Sabins and others
www.cell.com/cell-reports...
Really looking forward to see what plasmid aficionados think of this one!!
With @asantoslopez.bsky.social @wfigueroac3.bsky.social Akshay Sabins and others
www.cell.com/cell-reports...

Non-conjugative plasmids limit their mobility to persist in nature
Sabnis et al. explain why non-conjugative plasmids move at a low rate in nature. While
increased mobility can easily evolve by incorporating phage DNA into plasmids, this
is disadvantageous because it...
www.cell.com
October 22, 2025 at 1:12 PM
New paper with my (amazing) friend and mentor @jrpenades.bsky.social
Really looking forward to see what plasmid aficionados think of this one!!
With @asantoslopez.bsky.social @wfigueroac3.bsky.social Akshay Sabins and others
www.cell.com/cell-reports...
Really looking forward to see what plasmid aficionados think of this one!!
With @asantoslopez.bsky.social @wfigueroac3.bsky.social Akshay Sabins and others
www.cell.com/cell-reports...
Reposted by Jorge Sastre Domínguez
Our latest work reveals that arbitrium phages cross-communicate across species! These tiny viruses “listen” to signals from others, coordinating lysis-lysogeny decisions across species.
Original idea from @albertomarina.bsky.social and, as usual, he was right.
www.biorxiv.org/content/10.1...
Original idea from @albertomarina.bsky.social and, as usual, he was right.
www.biorxiv.org/content/10.1...

Phages communicate across species to shape microbial ecosystems
Arbitrium is a communication system that helps bacteriophages decide between lysis and lysogeny via secreted peptides. In arbitrium, the AimP peptide binds its cognate AimR receptor to repress aimX ex...
www.biorxiv.org
October 14, 2025 at 1:36 PM
Our latest work reveals that arbitrium phages cross-communicate across species! These tiny viruses “listen” to signals from others, coordinating lysis-lysogeny decisions across species.
Original idea from @albertomarina.bsky.social and, as usual, he was right.
www.biorxiv.org/content/10.1...
Original idea from @albertomarina.bsky.social and, as usual, he was right.
www.biorxiv.org/content/10.1...
Reposted by Jorge Sastre Domínguez
Can we exploit past phage infection events (prophages) to decipher the specificity of phage receptor-binding proteins such as depolymerases?🔎 Happy to share our recent work at @natcomms.nature.com 🔽 #microsky #phagesky
www.nature.com/articles/s41...
www.nature.com/articles/s41...

Unlocking data in Klebsiella lysogens to predict capsular type-specificity of phage depolymerases - Nature Communications
Here, the authors exploit the genetic information encoded in Klebsiella prophages to model the interplay between bacteria, prophages, and their depolymerases, using a directed acyclic graph-model and a sequence clustering-based model.
www.nature.com
October 8, 2025 at 8:59 AM
Can we exploit past phage infection events (prophages) to decipher the specificity of phage receptor-binding proteins such as depolymerases?🔎 Happy to share our recent work at @natcomms.nature.com 🔽 #microsky #phagesky
www.nature.com/articles/s41...
www.nature.com/articles/s41...
Reposted by Jorge Sastre Domínguez
Imagine we could travel back in time ⏪⌛️to explore the world of bacterial pathogens before humans discovered and industrialised antibiotics
We just did that to study the history of #AMR spread @science.org
doi.org/10.1126/scie...
If you like time travel & biology, this 🧵is for you👇
We just did that to study the history of #AMR spread @science.org
doi.org/10.1126/scie...
If you like time travel & biology, this 🧵is for you👇

Pre- and postantibiotic epoch: The historical spread of antimicrobial resistance
Plasmids are now the primary vectors of antimicrobial resistance, but our understanding of how human industrialisation of antibiotics influenced their evolution is limited by a paucity of data predati...
doi.org
October 6, 2025 at 10:42 AM
Imagine we could travel back in time ⏪⌛️to explore the world of bacterial pathogens before humans discovered and industrialised antibiotics
We just did that to study the history of #AMR spread @science.org
doi.org/10.1126/scie...
If you like time travel & biology, this 🧵is for you👇
We just did that to study the history of #AMR spread @science.org
doi.org/10.1126/scie...
If you like time travel & biology, this 🧵is for you👇
Reposted by Jorge Sastre Domínguez
Published in Current Biology! P. aeruginosa can use its filamentous phage to inhibit competitors but high phage production is susceptible to cheater miniphage invasion. Subsequent phage tragedy of the commons can lower bacteria and phage fitness. Link: authors.elsevier.com/c/1lt5I3QW8S...
Preprint out! Bacteria w/ hyper-replicative filamentous phage lead to overnight emergence of cheater phages. Bacteria w/ both phages can outcompete wildtype, then rapidly lose phage via a phage Tragedy of the Commons
@shellyscrib.bsky.social @vscooper.micropopbio.org www.biorxiv.org/content/10.1...
@shellyscrib.bsky.social @vscooper.micropopbio.org www.biorxiv.org/content/10.1...

Filamentous cheater phages drive bacterial and phage populations to lower fitness
Many bacteria carry phage genome(s) in their chromosome (i.e., prophage), and this intertwines the fitness of the bacterium and the phage. Most Pseudomonas aeruginosa strains carry filamentous phages ...
www.biorxiv.org
October 2, 2025 at 3:54 PM
Published in Current Biology! P. aeruginosa can use its filamentous phage to inhibit competitors but high phage production is susceptible to cheater miniphage invasion. Subsequent phage tragedy of the commons can lower bacteria and phage fitness. Link: authors.elsevier.com/c/1lt5I3QW8S...
Reposted by Jorge Sastre Domínguez
Precisely calling mutations across hundreds of bacterial isolates has been hard, requiring manual filtering and expertise.
Until now, using AccuSNV.
Herui Liao trained an ML model based on our previous meticulously called SNVs.
www.biorxiv.org/content/10.1...
Until now, using AccuSNV.
Herui Liao trained an ML model based on our previous meticulously called SNVs.
www.biorxiv.org/content/10.1...

High-accuracy SNV calling for bacterial isolates using deep learning with AccuSNV
Accurate detection of mutations within bacterial species is critical for fundamental studies of microbial evolution, reconstructing transmission events, and identifying antimicrobial resistance mutati...
www.biorxiv.org
September 29, 2025 at 7:45 PM
Precisely calling mutations across hundreds of bacterial isolates has been hard, requiring manual filtering and expertise.
Until now, using AccuSNV.
Herui Liao trained an ML model based on our previous meticulously called SNVs.
www.biorxiv.org/content/10.1...
Until now, using AccuSNV.
Herui Liao trained an ML model based on our previous meticulously called SNVs.
www.biorxiv.org/content/10.1...
Reposted by Jorge Sastre Domínguez
Delighted to see our paper studying the evolution of plasmids over the last 100 years, now out! Years of work by Adrian Cazares, also Nick Thomson @sangerinstitute.bsky.social - this version much improved over the preprint. Final version should be open access, apols.
Thread 1/n
Thread 1/n

September 25, 2025 at 9:29 PM
Delighted to see our paper studying the evolution of plasmids over the last 100 years, now out! Years of work by Adrian Cazares, also Nick Thomson @sangerinstitute.bsky.social - this version much improved over the preprint. Final version should be open access, apols.
Thread 1/n
Thread 1/n
Reposted by Jorge Sastre Domínguez
I am so proud to be part of this work, that we initiated Fernando de la Cruz and I, when he was on sabbatical in my lab in 2009... it took so much time for this achievement, 1000 thanks to Raúl Fernández-López! this brought me back to my PhD on cyanobacteria genetics. www.pnas.org/doi/10.1073/...

Mutations in the circadian cycle drive adaptive plasticity in cyanobacteria | PNAS
Circadian clocks allow organisms to anticipate daily fluctuations in light and temperature,
but how this anticipatory role promotes adaptation to d...
www.pnas.org
September 19, 2025 at 5:32 AM
I am so proud to be part of this work, that we initiated Fernando de la Cruz and I, when he was on sabbatical in my lab in 2009... it took so much time for this achievement, 1000 thanks to Raúl Fernández-López! this brought me back to my PhD on cyanobacteria genetics. www.pnas.org/doi/10.1073/...
Reposted by Jorge Sastre Domínguez
Sometimes you meet absolutely incredible bioinfo-magicians.
It was a huge privilege when @shenwei356.bsky.social
joined our group for a year on an @embl.org sabbatical.
While here, he developed a new way of aligning to
millions of bacteria, called LexicMap 1/n
www.nature.com/articles/s41...
It was a huge privilege when @shenwei356.bsky.social
joined our group for a year on an @embl.org sabbatical.
While here, he developed a new way of aligning to
millions of bacteria, called LexicMap 1/n
www.nature.com/articles/s41...

Efficient sequence alignment against millions of prokaryotic genomes with LexicMap - Nature Biotechnology
LexicMap uses a fixed set of probes to efficiently query gene sequences for fast and low-memory alignment.
www.nature.com
September 10, 2025 at 9:12 AM
Sometimes you meet absolutely incredible bioinfo-magicians.
It was a huge privilege when @shenwei356.bsky.social
joined our group for a year on an @embl.org sabbatical.
While here, he developed a new way of aligning to
millions of bacteria, called LexicMap 1/n
www.nature.com/articles/s41...
It was a huge privilege when @shenwei356.bsky.social
joined our group for a year on an @embl.org sabbatical.
While here, he developed a new way of aligning to
millions of bacteria, called LexicMap 1/n
www.nature.com/articles/s41...
Reposted by Jorge Sastre Domínguez
Really excited to share our latest paper led by @simonaube.bsky.social. Fascinating results examining whether regulatory mutations can lead to adaptation as fast as coding mutations do. #mevosky #evobio #evoSky
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

Adaptation is less accessible through mutations in promoters than in coding sequences when large effect sizes are needed
Mutations within promoters and coding sequences are both involved in adaptation, but their relative contributions remain to be compared directly. Using the fungal enzyme cytosine deaminase, we examine...
www.biorxiv.org
September 10, 2025 at 11:09 AM
Really excited to share our latest paper led by @simonaube.bsky.social. Fascinating results examining whether regulatory mutations can lead to adaptation as fast as coding mutations do. #mevosky #evobio #evoSky
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Reposted by Jorge Sastre Domínguez
Delighted to share our recently published work!
Ever wondered how Klebsiella (and others) deals with capsule production’s costs ?
The paper: journals.asm.org/doi/10.1128/...
Thread👇
Ever wondered how Klebsiella (and others) deals with capsule production’s costs ?
The paper: journals.asm.org/doi/10.1128/...
Thread👇

Phenotypic heterogeneity of capsule production across opportunistic pathogens | mBio
The polysaccharidic capsule is present in ~50% of species across the bacterial phylogeny, including all ESKAPE microorganisms, the six most significant multidrug-resistant (MDR) nosocomial pathogens. It is also an important virulence factor and a major ...
journals.asm.org
September 5, 2025 at 1:49 PM
Delighted to share our recently published work!
Ever wondered how Klebsiella (and others) deals with capsule production’s costs ?
The paper: journals.asm.org/doi/10.1128/...
Thread👇
Ever wondered how Klebsiella (and others) deals with capsule production’s costs ?
The paper: journals.asm.org/doi/10.1128/...
Thread👇
Reposted by Jorge Sastre Domínguez
Check out @julielebris.bsky.social’s thread on our latest manuscript describing phenotypic heterogeneity in capsule production in Klebsiella & Acinetobacter @klebclub.bsky.social
This work started when I was still in @pasteur.fr & got finished in @cbitoulouse.bsky.social
#microsky #phagesky
This work started when I was still in @pasteur.fr & got finished in @cbitoulouse.bsky.social
#microsky #phagesky
Delighted to share our recently published work!
Ever wondered how Klebsiella (and others) deals with capsule production’s costs ?
The paper: journals.asm.org/doi/10.1128/...
Thread👇
Ever wondered how Klebsiella (and others) deals with capsule production’s costs ?
The paper: journals.asm.org/doi/10.1128/...
Thread👇

Phenotypic heterogeneity of capsule production across opportunistic pathogens | mBio
The polysaccharidic capsule is present in ~50% of species across the bacterial phylogeny, including all ESKAPE microorganisms, the six most significant multidrug-resistant (MDR) nosocomial pathogens. It is also an important virulence factor and a major ...
journals.asm.org
September 9, 2025 at 5:11 AM
Check out @julielebris.bsky.social’s thread on our latest manuscript describing phenotypic heterogeneity in capsule production in Klebsiella & Acinetobacter @klebclub.bsky.social
This work started when I was still in @pasteur.fr & got finished in @cbitoulouse.bsky.social
#microsky #phagesky
This work started when I was still in @pasteur.fr & got finished in @cbitoulouse.bsky.social
#microsky #phagesky
Reposted by Jorge Sastre Domínguez
We are hiring a PhD student! Are you fascinated by microbes and evolution? Come join us in Barcelona!
#WeAreHiring
📣 The #IBE_Barcelona is seeking a #PhD Student for the Evolutionary #Microbiology Lab.🔬🧫
⏳ Apply before September 30th and #JoinOurTeam!
#UnravellingEvolution #ConservingBiodiversity
✍ https://www.ibe.upf-csic.es/work-with-us/job-offers
📣 The #IBE_Barcelona is seeking a #PhD Student for the Evolutionary #Microbiology Lab.🔬🧫
⏳ Apply before September 30th and #JoinOurTeam!
#UnravellingEvolution #ConservingBiodiversity
✍ https://www.ibe.upf-csic.es/work-with-us/job-offers

September 10, 2025 at 8:39 AM
We are hiring a PhD student! Are you fascinated by microbes and evolution? Come join us in Barcelona!
Reposted by Jorge Sastre Domínguez
How complex functions, with important physiological and evolutionary impacts get repeatedly and efficiently transferred across genomes?
That’s what we explored using one of the fastest-evolving loci in Bacteria: the capsule locus.
The paper: www.biorxiv.org/content/10.1...
Thread👇
That’s what we explored using one of the fastest-evolving loci in Bacteria: the capsule locus.
The paper: www.biorxiv.org/content/10.1...
Thread👇

Serotype swapping in Klebsiella spp. by plug-and-play
Understanding how complex, multi-gene systems evolve and function across genetic backgrounds is a central question in molecular evolution. While such systems often impose costs through epistatic inter...
www.biorxiv.org
September 10, 2025 at 8:17 AM
How complex functions, with important physiological and evolutionary impacts get repeatedly and efficiently transferred across genomes?
That’s what we explored using one of the fastest-evolving loci in Bacteria: the capsule locus.
The paper: www.biorxiv.org/content/10.1...
Thread👇
That’s what we explored using one of the fastest-evolving loci in Bacteria: the capsule locus.
The paper: www.biorxiv.org/content/10.1...
Thread👇
Reposted by Jorge Sastre Domínguez
For anyone who has used pling for comparing plasmids using rearrangement distances ("how many structural events apart are these plasmids"), here's how to tweak parameters, and integrate it with typing info, and the host phylogeny
www.biorxiv.org/content/10.1...
github.com/iqbal-lab-or...
www.biorxiv.org/content/10.1...
github.com/iqbal-lab-or...

Clustering of plasmid genomes for genomic epidemiology by using rearrangement distances, with pling
Integration of plasmids into genomic epidemiology is challenging, because there are no clearly defined evolving-units (equivalent to species), and because plasmids appear to evolve as much by structur...
www.biorxiv.org
September 7, 2025 at 2:56 PM
For anyone who has used pling for comparing plasmids using rearrangement distances ("how many structural events apart are these plasmids"), here's how to tweak parameters, and integrate it with typing info, and the host phylogeny
www.biorxiv.org/content/10.1...
github.com/iqbal-lab-or...
www.biorxiv.org/content/10.1...
github.com/iqbal-lab-or...
Reposted by Jorge Sastre Domínguez
El grupo de Álvaro San Millán revela el coste de mantener la resistencia en enterobacterias
El trabajo traza un mapa funcional del plásmido pOXA-48, con potencial para guiar nuevas terapias contra resistencias
bit.ly/4p0fCmY
@sanmillan.bsky.social @aliciapcv.bsky.social @jorgesastred.bsky.social
El trabajo traza un mapa funcional del plásmido pOXA-48, con potencial para guiar nuevas terapias contra resistencias
bit.ly/4p0fCmY
@sanmillan.bsky.social @aliciapcv.bsky.social @jorgesastred.bsky.social

Identifican el coste fisiológico de mantener la resistencia microbiana en enterobacterias - CNB
bit.ly
August 27, 2025 at 8:32 AM
El grupo de Álvaro San Millán revela el coste de mantener la resistencia en enterobacterias
El trabajo traza un mapa funcional del plásmido pOXA-48, con potencial para guiar nuevas terapias contra resistencias
bit.ly/4p0fCmY
@sanmillan.bsky.social @aliciapcv.bsky.social @jorgesastred.bsky.social
El trabajo traza un mapa funcional del plásmido pOXA-48, con potencial para guiar nuevas terapias contra resistencias
bit.ly/4p0fCmY
@sanmillan.bsky.social @aliciapcv.bsky.social @jorgesastred.bsky.social
Extremely glad to see this out!!
Wonderful work by all the lab, especially @aliciapcv.bsky.social. Don't miss her thread!!
Wonderful work by all the lab, especially @aliciapcv.bsky.social. Don't miss her thread!!
This work is finally published! 🥳🧬
Plasmids are associated with very variable fitness costs in their different bacterial hosts. But, what is the contribution of each of the plasmid-genes in these host-specific effects? Study led by
@jorgesastred.bsky.social, @sanmillan.bsky.social and myself! 1/14
Plasmids are associated with very variable fitness costs in their different bacterial hosts. But, what is the contribution of each of the plasmid-genes in these host-specific effects? Study led by
@jorgesastred.bsky.social, @sanmillan.bsky.social and myself! 1/14
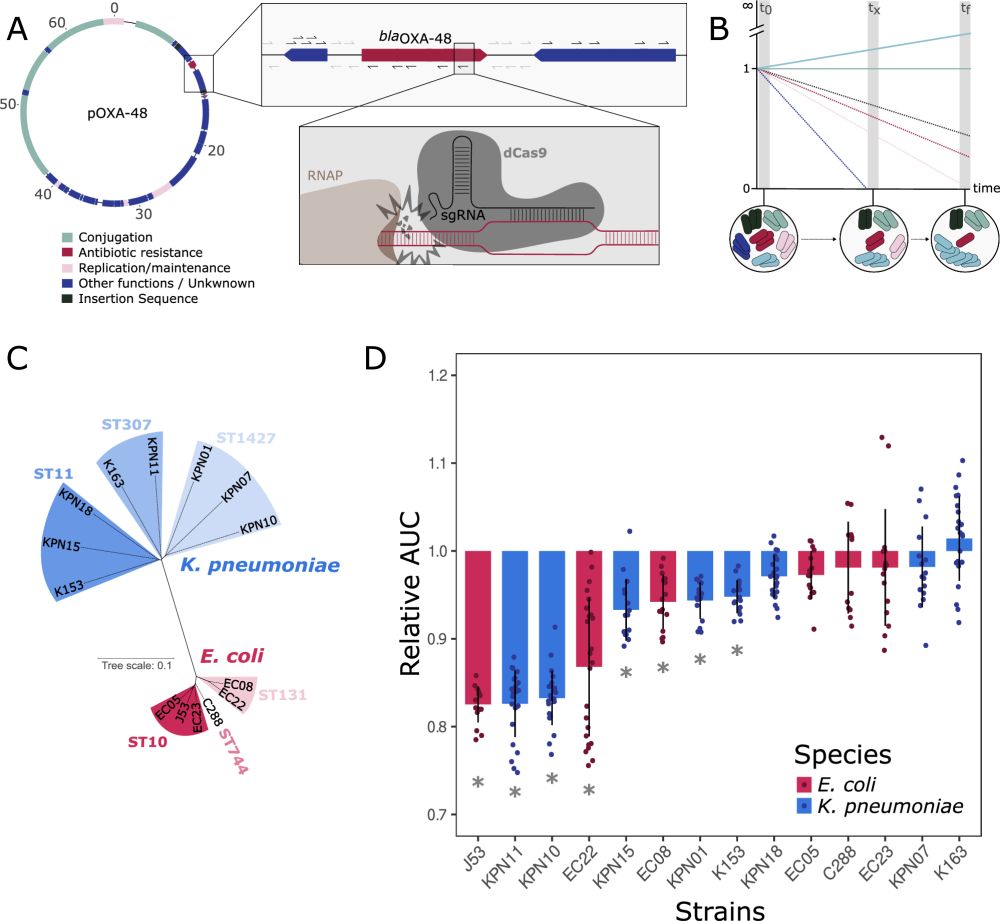
Dissecting pOXA-48 fitness effects in clinical Enterobacterales using plasmid-wide CRISPRi screens
Nature Communications - This study investigates the effects of the carbapenem resistance plasmid pOXA-48 in clinical enterobacteria. Using CRISPRi screens, the authors revealed that the...
rdcu.be
August 22, 2025 at 9:17 AM
Extremely glad to see this out!!
Wonderful work by all the lab, especially @aliciapcv.bsky.social. Don't miss her thread!!
Wonderful work by all the lab, especially @aliciapcv.bsky.social. Don't miss her thread!!
Reposted by Jorge Sastre Domínguez
Our new manuscript is out! A bit of everything cool:
Plasmids ✅
Insertion Sequences ✅
AMR Evolution ✅
Microbial Communities ✅
Databases analyses ✅
Mathematical modeling ✅
See the scientific thread below of Jorge Sastre, who has brilliantly led this work with @palomarodera.bsky.social
Plasmids ✅
Insertion Sequences ✅
AMR Evolution ✅
Microbial Communities ✅
Databases analyses ✅
Mathematical modeling ✅
See the scientific thread below of Jorge Sastre, who has brilliantly led this work with @palomarodera.bsky.social
New paper out! 🔈🔈📣📣
Plasmids promote antimicrobial resistance through Insertion Sequence-mediated gene inactivation.
Combining experimental and computational approaches, we unveil how two of the most prevalent bacterial MGE accelerate the evolution of AMR. 🧵👇🏻
www.biorxiv.org/content/10.1...
Plasmids promote antimicrobial resistance through Insertion Sequence-mediated gene inactivation.
Combining experimental and computational approaches, we unveil how two of the most prevalent bacterial MGE accelerate the evolution of AMR. 🧵👇🏻
www.biorxiv.org/content/10.1...

Plasmids promote antimicrobial resistance through Insertion Sequence-mediated gene inactivation
Antimicrobial Resistance (AMR) is a major threat to public health. Plasmids are mobile genetic elements that can rapidly spread across bacterial populations, promoting the dissemination of AMR genes i...
www.biorxiv.org
August 13, 2025 at 3:04 PM
Our new manuscript is out! A bit of everything cool:
Plasmids ✅
Insertion Sequences ✅
AMR Evolution ✅
Microbial Communities ✅
Databases analyses ✅
Mathematical modeling ✅
See the scientific thread below of Jorge Sastre, who has brilliantly led this work with @palomarodera.bsky.social
Plasmids ✅
Insertion Sequences ✅
AMR Evolution ✅
Microbial Communities ✅
Databases analyses ✅
Mathematical modeling ✅
See the scientific thread below of Jorge Sastre, who has brilliantly led this work with @palomarodera.bsky.social
Reposted by Jorge Sastre Domínguez
If you ever find yourself needing evidence for ‘Plasmids are just as common in microbes without resistance genes,’ we’ve got you covered! Check our new paper, out today:
www.microbiologyresearch.org/content/jour...
www.microbiologyresearch.org/content/jour...

Plasmid prevalence is independent of antibiotic resistance in environmental Enterobacteriaceae
The rapid rise of antibiotic-resistant pathogens poses a critical threat to the treatment of infectious diseases. While the spread of antibiotic resistance genes (ARGs) via plasmid conjugation has bee...
www.microbiologyresearch.org
August 12, 2025 at 5:14 PM
If you ever find yourself needing evidence for ‘Plasmids are just as common in microbes without resistance genes,’ we’ve got you covered! Check our new paper, out today:
www.microbiologyresearch.org/content/jour...
www.microbiologyresearch.org/content/jour...
Reposted by Jorge Sastre Domínguez
August 13, 2025 at 6:39 AM


