
Congratultions to @lizebonaaugust.bsky.social Blanca del Rosal and Leslie Yeo it was a pleasure to work together on this amazing project! 🎊🎊🎊🎊

Congratultions to @lizebonaaugust.bsky.social Blanca del Rosal and Leslie Yeo it was a pleasure to work together on this amazing project! 🎊🎊🎊🎊
To join, please follow this link: qmul.ac.uk/epigenetics/...

register here for the entire series: us06web.zoom.us/webinar/regi...

register here for the entire series: us06web.zoom.us/webinar/regi...

We find the rate of cohesin loop extrusion in cells is set by NIPBL dosage and tunes many aspects of chromosome folding.
This provides a molecular basis for NIPBL haploinsufficiency in humans. 🧵👇
www.biorxiv.org/content/10.1...

We find the rate of cohesin loop extrusion in cells is set by NIPBL dosage and tunes many aspects of chromosome folding.
This provides a molecular basis for NIPBL haploinsufficiency in humans. 🧵👇
www.biorxiv.org/content/10.1...
To join, please follow this link: qmul.ac.uk/epigenetics/...

To join, please follow this link: qmul.ac.uk/epigenetics/...
📰 edin.ac/3H1qnE1...




Latest paper from our fruitful collaboration with Emily Rocha and Tim Greenamyre, linking epigenetic signatures of exposure to toxicants to Parkinson's disease:
www.nature.com/articles/s41...
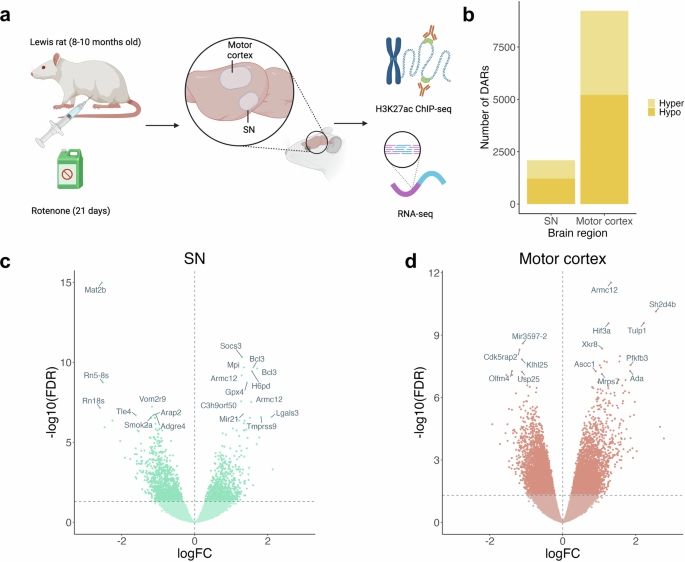
Latest paper from our fruitful collaboration with Emily Rocha and Tim Greenamyre, linking epigenetic signatures of exposure to toxicants to Parkinson's disease:
www.nature.com/articles/s41...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
that explores how PRC1-mediated H2AK119ub deposition safeguards lineage fidelity during forebrain development. Fantastic work by @lucyadoyle.bsky.social and our colleagues & collaborators. 🥳🧬🧠🔬
www.biorxiv.org/content/10.1...

that explores how PRC1-mediated H2AK119ub deposition safeguards lineage fidelity during forebrain development. Fantastic work by @lucyadoyle.bsky.social and our colleagues & collaborators. 🥳🧬🧠🔬
www.biorxiv.org/content/10.1...

