@QMULepigenetics, views are my own.
If you want to learn how targeting cohesin to defined loci in the genome affects the local chromatin environment and transcription, look no further!
rdcu.be/eLiT5

If you want to learn how targeting cohesin to defined loci in the genome affects the local chromatin environment and transcription, look no further!
rdcu.be/eLiT5



In a new preprint, we share some clues about when, how, and why this happens!
www.biorxiv.org/content/10.1...

In a new preprint, we share some clues about when, how, and why this happens!
www.biorxiv.org/content/10.1...


To join, please follow this link: qmul.ac.uk/epigenetics/...



@imperiallifesci.bsky.social
@imperiallifesci.bsky.social
To join, please follow this link: qmul.ac.uk/epigenetics/...

To join, please follow this link: qmul.ac.uk/epigenetics/...
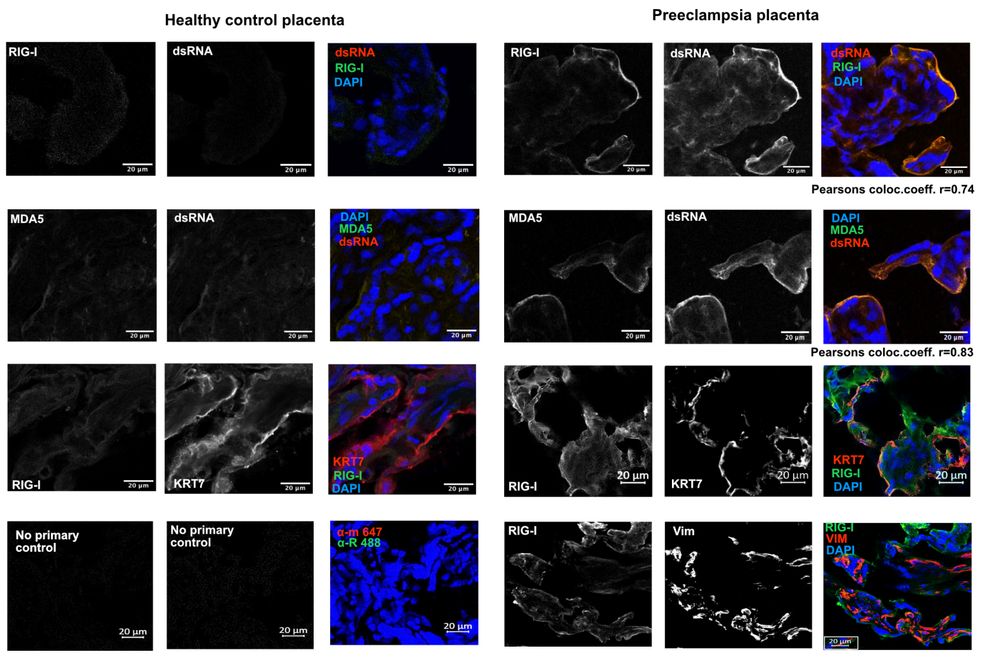
But how can just one histone modification (H3K27ac) distinguish an enhancer from a non-enhancer?
While still a valuable indicator, it is just one component of a complex feature.
Great paper and very important finding - many congrats Axel & team! 👏👏
But how can just one histone modification (H3K27ac) distinguish an enhancer from a non-enhancer?
While still a valuable indicator, it is just one component of a complex feature.
I am excited to share our new work on epigenetic enhancer priming in early mammalian development
genomebiology.biomedcentral.com/articles/10....
Fantastic collaboration with Wolf Reik’s team, with key contributions from many others, led by the excellent Chris Todd.
Short 🧵
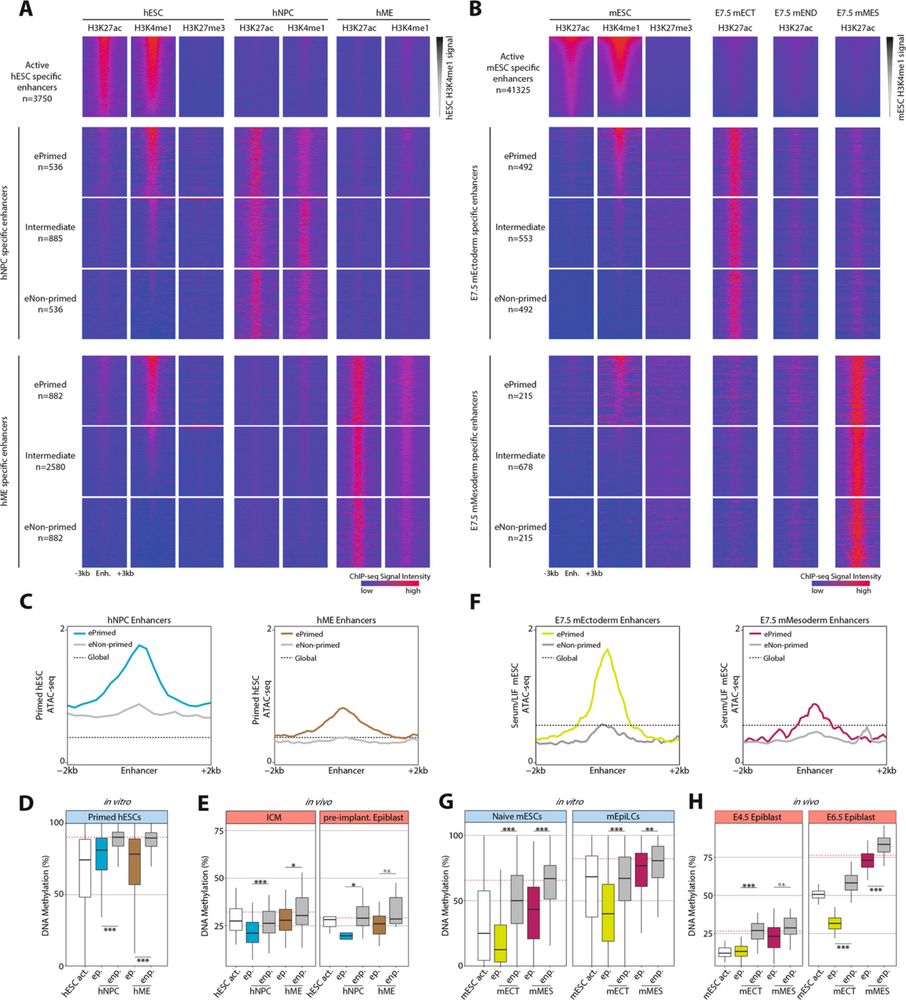
I am excited to share our new work on epigenetic enhancer priming in early mammalian development
genomebiology.biomedcentral.com/articles/10....
Fantastic collaboration with Wolf Reik’s team, with key contributions from many others, led by the excellent Chris Todd.
Short 🧵
that explores how PRC1-mediated H2AK119ub deposition safeguards lineage fidelity during forebrain development. Fantastic work by @lucyadoyle.bsky.social and our colleagues & collaborators. 🥳🧬🧠🔬
www.biorxiv.org/content/10.1...

that explores how PRC1-mediated H2AK119ub deposition safeguards lineage fidelity during forebrain development. Fantastic work by @lucyadoyle.bsky.social and our colleagues & collaborators. 🥳🧬🧠🔬
www.biorxiv.org/content/10.1...
Please register as soon as possible and submit abstracts for oral presentations and posters.

Please register as soon as possible and submit abstracts for oral presentations and posters.
Pl register ow.ly/7QQb50W9Alo & share

Pl register ow.ly/7QQb50W9Alo & share
Bas Heijmans and @elisaoricchio.bsky.social for interaction with our centre.


In collaboration with @herassr.bsky.social and @heick.bsky.social labs
www.biorxiv.org/content/10.1...

In collaboration with @herassr.bsky.social and @heick.bsky.social labs
www.biorxiv.org/content/10.1...
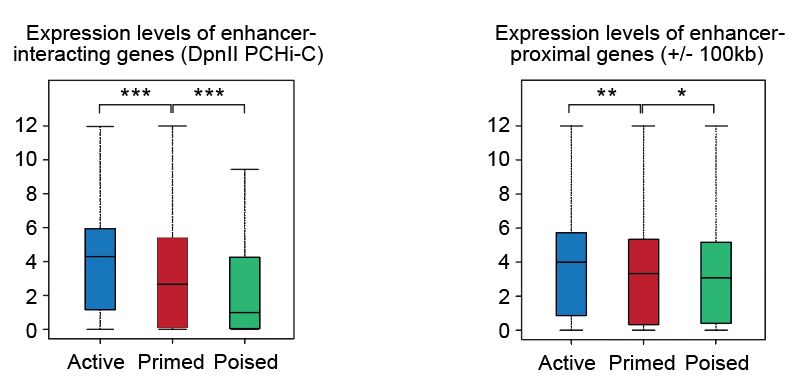
- Active (ATAC-seq+; H3K4me1+; H3K27ac+)
- Primed/Inactive (ATAC-seq+; H3K4me1+; H3K27ac-)
- Poised (ATAC-seq+; H3K27me3+)
- Chromatin repressed (ATAC-seq-)

- Active (ATAC-seq+; H3K4me1+; H3K27ac+)
- Primed/Inactive (ATAC-seq+; H3K4me1+; H3K27ac-)
- Poised (ATAC-seq+; H3K27me3+)
- Chromatin repressed (ATAC-seq-)
www.biorxiv.org/content/10.1...
🧵 below

From the next application rounds, expect changes to the:
• proposal structure
• evaluation process
• extra funding you can request
• eligibility for Starting & Consolidator #Grants (from 2027)
More 👇 europa.eu/!RPHWvv


