
Gregor Sturm
@grst.bsky.social
Single Cell/Spatial. Cancer Immunology. Outdoor activities.
Core developer @scverse.bsky.social.
Working in Clinical Bioinformatics at Boehringer Ingelheim.
Formerly PhD student at Medical University of Innsbruck.
My private account.
github.com/grst
Core developer @scverse.bsky.social.
Working in Clinical Bioinformatics at Boehringer Ingelheim.
Formerly PhD student at Medical University of Innsbruck.
My private account.
github.com/grst
There's another scverse conference this year and it will be amazing!
Register now: www.eventbrite.com/e/scverse-co...
Register now: www.eventbrite.com/e/scverse-co...
🎉 scverse conference 2025 Registration & Call for Abstracts NOW OPEN! 🎉
We're excited to announce that registration and the call for abstracts are officially open for the scverse Conference 2025!
Details in thread!
🧵 1/3
We're excited to announce that registration and the call for abstracts are officially open for the scverse Conference 2025!
Details in thread!
🧵 1/3
August 22, 2025 at 5:56 AM
There's another scverse conference this year and it will be amazing!
Register now: www.eventbrite.com/e/scverse-co...
Register now: www.eventbrite.com/e/scverse-co...
Reposted by Gregor Sturm
🎉 scverse conference 2025 Registration & Call for Abstracts NOW OPEN! 🎉
We're excited to announce that registration and the call for abstracts are officially open for the scverse Conference 2025!
Details in thread!
🧵 1/3
We're excited to announce that registration and the call for abstracts are officially open for the scverse Conference 2025!
Details in thread!
🧵 1/3
August 2, 2025 at 3:00 PM
🎉 scverse conference 2025 Registration & Call for Abstracts NOW OPEN! 🎉
We're excited to announce that registration and the call for abstracts are officially open for the scverse Conference 2025!
Details in thread!
🧵 1/3
We're excited to announce that registration and the call for abstracts are officially open for the scverse Conference 2025!
Details in thread!
🧵 1/3
Our benchmark + guidelines for atlas-level differential gene expression of single cells is online:
academic.oup.com/bib/article/...
Bottom line: Use pseudobulk + DESeq2 in simple and pseudobulk + DREAM in more complex settings.
Collab w/ @leonhafner.bsky.social @itisalist.bsky.social
academic.oup.com/bib/article/...
Bottom line: Use pseudobulk + DESeq2 in simple and pseudobulk + DREAM in more complex settings.
Collab w/ @leonhafner.bsky.social @itisalist.bsky.social

August 13, 2025 at 5:51 AM
Our benchmark + guidelines for atlas-level differential gene expression of single cells is online:
academic.oup.com/bib/article/...
Bottom line: Use pseudobulk + DESeq2 in simple and pseudobulk + DREAM in more complex settings.
Collab w/ @leonhafner.bsky.social @itisalist.bsky.social
academic.oup.com/bib/article/...
Bottom line: Use pseudobulk + DESeq2 in simple and pseudobulk + DREAM in more complex settings.
Collab w/ @leonhafner.bsky.social @itisalist.bsky.social
Register now for the best conference of the year!
🎉 scverse conference 2025 Registration & Call for Abstracts NOW OPEN! 🎉
We're excited to announce that registration and the call for abstracts are officially open for the scverse Conference 2025!
Details in thread!
🧵 1/3
We're excited to announce that registration and the call for abstracts are officially open for the scverse Conference 2025!
Details in thread!
🧵 1/3
August 5, 2025 at 5:47 AM
Register now for the best conference of the year!
Reposted by Gregor Sturm
📣 Mark your calendars! The 2025 edition of the scverse conference will take place on 17-19 November at Stanford University (US) scverse.org/conference20...
Call for abstracts and registrations coming soon!
Call for abstracts and registrations coming soon!

scverse conference 2025
Follow us on our channels to learn more details in the coming weeks
scverse.org
May 12, 2025 at 10:47 PM
📣 Mark your calendars! The 2025 edition of the scverse conference will take place on 17-19 November at Stanford University (US) scverse.org/conference20...
Call for abstracts and registrations coming soon!
Call for abstracts and registrations coming soon!
Just released a new version of the @scverse.bsky.social cookiecutter template: github.com/scverse/cook...
Some highlights:
🔃 improved template sync (merge conflicts now show up as such)
🚀 use hatch as project manager
🔧 lots of fixes and documentation updates
Some highlights:
🔃 improved template sync (merge conflicts now show up as such)
🚀 use hatch as project manager
🔧 lots of fixes and documentation updates
Release v0.5.0 · scverse/cookiecutter-scverse
New template sync
We re-implemented template sync from scratch instead on relying on cruft. This allows us to create real merge conflicts that show up as such on GitHub instead of .rej files.
Gene...
github.com
April 2, 2025 at 6:37 PM
Just released a new version of the @scverse.bsky.social cookiecutter template: github.com/scverse/cook...
Some highlights:
🔃 improved template sync (merge conflicts now show up as such)
🚀 use hatch as project manager
🔧 lots of fixes and documentation updates
Some highlights:
🔃 improved template sync (merge conflicts now show up as such)
🚀 use hatch as project manager
🔧 lots of fixes and documentation updates
Reposted by Gregor Sturm
Rogue Scholar
rogue-scholar.org
March 14, 2025 at 10:01 AM
Reposted by Gregor Sturm
Blog post by @const-ae.bsky.social with a simple explanation of the manifold regression algorithm & code that underlies our paper “Analysis of multi-condition single-cell data with latent embedding multivariate regression” (doi.org/10.1002/eji....).
const-ae.name/post/2025-01...
const-ae.name/post/2025-01...

LEMUR simplified | const-ae
A simplified implementation of the LEMUR algorithm.
const-ae.name
March 4, 2025 at 6:27 PM
Blog post by @const-ae.bsky.social with a simple explanation of the manifold regression algorithm & code that underlies our paper “Analysis of multi-condition single-cell data with latent embedding multivariate regression” (doi.org/10.1002/eji....).
const-ae.name/post/2025-01...
const-ae.name/post/2025-01...
Just released scirpy v0.21 -- Now with GPU Support for Hamming sequence distance and a brand new tutorial for working with scTCR datasets >1M cells: scirpy.scverse.org/en/latest/tu...
@scverse.bsky.social
@scverse.bsky.social
Working with >1M cells
Scirpy scales to millions of cells on a single workstation. This page is a work-in-progess collection with advice how to work with large datasets. Distance metrics: Computing pairwise sequence dist...
scirpy.scverse.org
February 25, 2025 at 5:46 PM
Just released scirpy v0.21 -- Now with GPU Support for Hamming sequence distance and a brand new tutorial for working with scTCR datasets >1M cells: scirpy.scverse.org/en/latest/tu...
@scverse.bsky.social
@scverse.bsky.social
Reposted by Gregor Sturm
🎉 Scanpy 1.11.0 is out! 🎉 just after reaching 2000 stars on GitHub!
- sc.pp.sample replaces subsample with many new features
- Sparse Dask support pca
- session-info2 package for more reproducible notebooks
See the release notes:
- sc.pp.sample replaces subsample with many new features
- Sparse Dask support pca
- session-info2 package for more reproducible notebooks
See the release notes:

Release notes
Version 1.11: 1.11.0 2025-02-14: Release candidates: rc2 2025-01-24, rc1 2024-12-20. Features: rc1 sample() supports both upsampling and downsampling of observations and variables. subsample() is n...
buff.ly
February 14, 2025 at 12:08 PM
🎉 Scanpy 1.11.0 is out! 🎉 just after reaching 2000 stars on GitHub!
- sc.pp.sample replaces subsample with many new features
- Sparse Dask support pca
- session-info2 package for more reproducible notebooks
See the release notes:
- sc.pp.sample replaces subsample with many new features
- Sparse Dask support pca
- session-info2 package for more reproducible notebooks
See the release notes:
Reposted by Gregor Sturm
Been looking forward to this talk since @alexpeltzer.bsky.social told me about DSO in October!
If you want to learn more, I'll be presenting this at a @nf-co.re bytesize talk: nf-co.re/events/2025/...

Bytesize: data science operations (DSO)
Gregor Sturm, Boehringer Ingelheim
nf-co.re
February 9, 2025 at 2:07 PM
Been looking forward to this talk since @alexpeltzer.bsky.social told me about DSO in October!
Reposted by Gregor Sturm
I'd like to share DSO, a command line helper to build reproducible data science projects with ease.
It is an opinionated way to organize data science projects, built around data version control (DVC).
github.com/Boehringer-I...
It is an opinionated way to organize data science projects, built around data version control (DVC).
github.com/Boehringer-I...

GitHub - Boehringer-Ingelheim/dso: Data Science Operations (dso) command line tool
Data Science Operations (dso) command line tool. Contribute to Boehringer-Ingelheim/dso development by creating an account on GitHub.
github.com
February 5, 2025 at 6:32 PM
I'd like to share DSO, a command line helper to build reproducible data science projects with ease.
It is an opinionated way to organize data science projects, built around data version control (DVC).
github.com/Boehringer-I...
It is an opinionated way to organize data science projects, built around data version control (DVC).
github.com/Boehringer-I...
I'd like to share DSO, a command line helper to build reproducible data science projects with ease.
It is an opinionated way to organize data science projects, built around data version control (DVC).
github.com/Boehringer-I...
It is an opinionated way to organize data science projects, built around data version control (DVC).
github.com/Boehringer-I...

GitHub - Boehringer-Ingelheim/dso: Data Science Operations (dso) command line tool
Data Science Operations (dso) command line tool. Contribute to Boehringer-Ingelheim/dso development by creating an account on GitHub.
github.com
February 5, 2025 at 6:32 PM
I'd like to share DSO, a command line helper to build reproducible data science projects with ease.
It is an opinionated way to organize data science projects, built around data version control (DVC).
github.com/Boehringer-I...
It is an opinionated way to organize data science projects, built around data version control (DVC).
github.com/Boehringer-I...
Reposted by Gregor Sturm
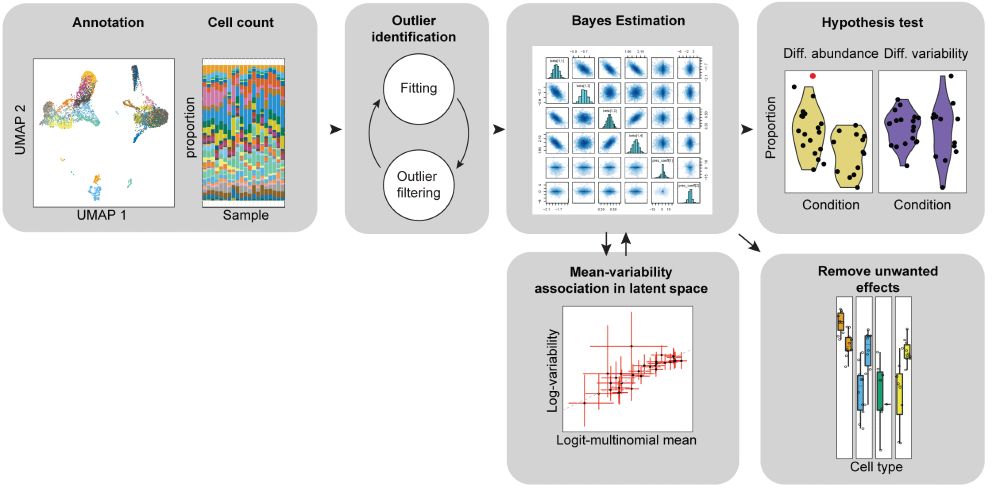
We (Chen Zhan!) just launched #sccomp for #Python!
Testing for differences in cell-type proportion in #singlecell #spatial data?
#sccomp is a mixed-effect Bayesian model
- Use sum-constrained BetaBinomial distribution
- Outliers detect.
- Remove unwanted effects
github.com/MangiolaLabo...
Testing for differences in cell-type proportion in #singlecell #spatial data?
#sccomp is a mixed-effect Bayesian model
- Use sum-constrained BetaBinomial distribution
- Outliers detect.
- Remove unwanted effects
github.com/MangiolaLabo...
January 22, 2025 at 2:58 AM
We (Chen Zhan!) just launched #sccomp for #Python!
Testing for differences in cell-type proportion in #singlecell #spatial data?
#sccomp is a mixed-effect Bayesian model
- Use sum-constrained BetaBinomial distribution
- Outliers detect.
- Remove unwanted effects
github.com/MangiolaLabo...
Testing for differences in cell-type proportion in #singlecell #spatial data?
#sccomp is a mixed-effect Bayesian model
- Use sum-constrained BetaBinomial distribution
- Outliers detect.
- Remove unwanted effects
github.com/MangiolaLabo...
Two years after publication of our single-cell lung cancer atlas, a user found a mistake in the annotation of the EGFR-status of some patients. We fixed the issue and the atlas is now updated on cell-x-gene: cellxgene.cziscience.com/collections/...
What are the takeaways from that? (1/3)
What are the takeaways from that? (1/3)
Cellxgene Data Portal
Find, download, and visually explore curated and standardized single cell datasets.
cellxgene.cziscience.com
January 19, 2025 at 10:54 AM
Two years after publication of our single-cell lung cancer atlas, a user found a mistake in the annotation of the EGFR-status of some patients. We fixed the issue and the atlas is now updated on cell-x-gene: cellxgene.cziscience.com/collections/...
What are the takeaways from that? (1/3)
What are the takeaways from that? (1/3)
Reposted by Gregor Sturm
I am Stoked about our upcoming @scverse.bsky.social
and @owkin.bsky.social hackathon, focused on spatial omics data analysis.
📅 March 17-19, 2025
📍 Owkin office, Paris
Apply now: docs.google.com/forms/d/e/1F...
and @owkin.bsky.social hackathon, focused on spatial omics data analysis.
📅 March 17-19, 2025
📍 Owkin office, Paris
Apply now: docs.google.com/forms/d/e/1F...

Scverse x Owkin Hackathon in Paris
We're pleased to announce the next Scverse Hackathon will take place in the Owkin offices in Paris from 17/03/2025 9am to 19/03/2025 1:30pm.
This hackathon is a joint initiative between the scverse c...
docs.google.com
January 17, 2025 at 10:34 AM
I am Stoked about our upcoming @scverse.bsky.social
and @owkin.bsky.social hackathon, focused on spatial omics data analysis.
📅 March 17-19, 2025
📍 Owkin office, Paris
Apply now: docs.google.com/forms/d/e/1F...
and @owkin.bsky.social hackathon, focused on spatial omics data analysis.
📅 March 17-19, 2025
📍 Owkin office, Paris
Apply now: docs.google.com/forms/d/e/1F...
protein sequencing 👀
1/ We’re excited to announce our first speaker of London Calling 2025 Jeff Nivala will share their groundbreaking ‘proof-of-concept' method showcasing the capability of #nanopore sequencing to read #polypeptide strands.
Register here: nanoporetech.com/about/events... #nanoporeconf
Register here: nanoporetech.com/about/events... #nanoporeconf

January 14, 2025 at 7:53 PM
protein sequencing 👀
Reposted by Gregor Sturm
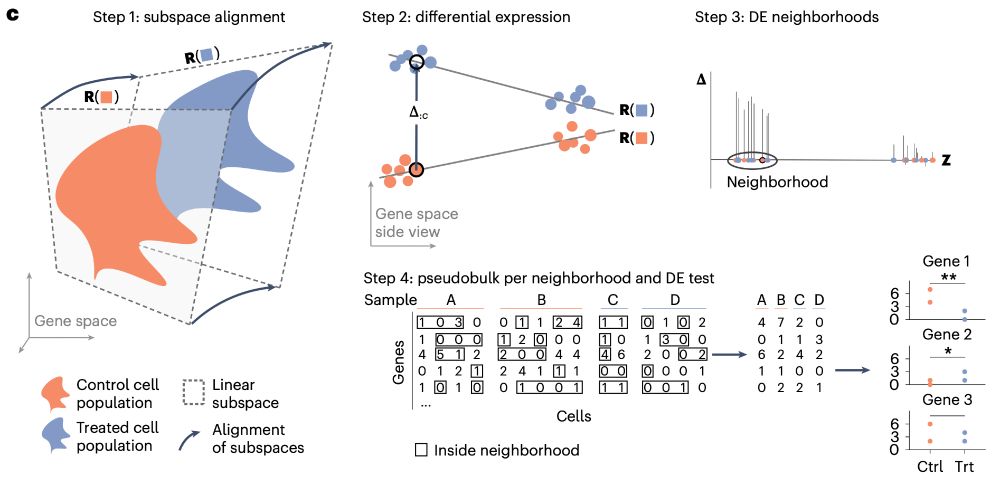
After 4y in the making, I am super excited that my main PhD project is published 🎉🥳🎉🎉🥳
www.nature.com/articles/s41...
LEMUR is a tool to analyze multi-condition single-cell data and model differential expression as a continuous function of the cell-state space.
Some highlights⬇️
www.nature.com/articles/s41...
LEMUR is a tool to analyze multi-condition single-cell data and model differential expression as a continuous function of the cell-state space.
Some highlights⬇️
January 3, 2025 at 7:25 PM
After 4y in the making, I am super excited that my main PhD project is published 🎉🥳🎉🎉🥳
www.nature.com/articles/s41...
LEMUR is a tool to analyze multi-condition single-cell data and model differential expression as a continuous function of the cell-state space.
Some highlights⬇️
www.nature.com/articles/s41...
LEMUR is a tool to analyze multi-condition single-cell data and model differential expression as a continuous function of the cell-state space.
Some highlights⬇️
Dynamic electricity tariffs are an incentive to use energy when it's abundant and emits little CO2.
But are they also cheaper?
✅ For 2024, we would have saved 10-13% compared to our current fixed price tariff. Without any optimization.
Full post (in German): grst.github.io/dynamischer-...
But are they also cheaper?
✅ For 2024, we would have saved 10-13% compared to our current fixed price tariff. Without any optimization.
Full post (in German): grst.github.io/dynamischer-...
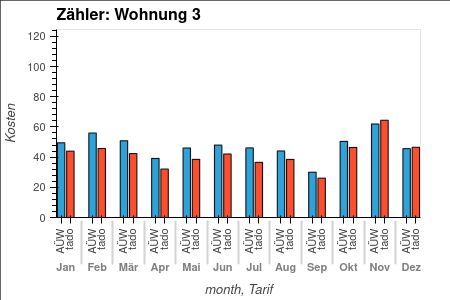
January 2, 2025 at 1:35 PM
Dynamic electricity tariffs are an incentive to use energy when it's abundant and emits little CO2.
But are they also cheaper?
✅ For 2024, we would have saved 10-13% compared to our current fixed price tariff. Without any optimization.
Full post (in German): grst.github.io/dynamischer-...
But are they also cheaper?
✅ For 2024, we would have saved 10-13% compared to our current fixed price tariff. Without any optimization.
Full post (in German): grst.github.io/dynamischer-...
Reposted by Gregor Sturm
To bring to light data science topics that usually don’t make it into publications I started a blog on this topic: hrovatin.github.io By interviewing different researchers, I plan to find out what is going on in the community.
Karin Hrovatin
Data science blog on topics that don’t get published.
hrovatin.github.io
December 16, 2024 at 6:59 AM
To bring to light data science topics that usually don’t make it into publications I started a blog on this topic: hrovatin.github.io By interviewing different researchers, I plan to find out what is going on in the community.
Reposted by Gregor Sturm
Formulaic is the go-to way to specify design formulas in Python, e.g. ~treatment + timepoint.
To compare sth, one needs to specify a contrast, e.g "on treatment vs baseline".
To make this easier, we developed "formulaic-contrasts":
formulaic-contrasts.readthedocs.io/en/latest/
To compare sth, one needs to specify a contrast, e.g "on treatment vs baseline".
To make this easier, we developed "formulaic-contrasts":
formulaic-contrasts.readthedocs.io/en/latest/

December 7, 2024 at 7:05 PM
Formulaic is the go-to way to specify design formulas in Python, e.g. ~treatment + timepoint.
To compare sth, one needs to specify a contrast, e.g "on treatment vs baseline".
To make this easier, we developed "formulaic-contrasts":
formulaic-contrasts.readthedocs.io/en/latest/
To compare sth, one needs to specify a contrast, e.g "on treatment vs baseline".
To make this easier, we developed "formulaic-contrasts":
formulaic-contrasts.readthedocs.io/en/latest/
Formulaic is the go-to way to specify design formulas in Python, e.g. ~treatment + timepoint.
To compare sth, one needs to specify a contrast, e.g "on treatment vs baseline".
To make this easier, we developed "formulaic-contrasts":
formulaic-contrasts.readthedocs.io/en/latest/
To compare sth, one needs to specify a contrast, e.g "on treatment vs baseline".
To make this easier, we developed "formulaic-contrasts":
formulaic-contrasts.readthedocs.io/en/latest/

December 7, 2024 at 7:05 PM
Formulaic is the go-to way to specify design formulas in Python, e.g. ~treatment + timepoint.
To compare sth, one needs to specify a contrast, e.g "on treatment vs baseline".
To make this easier, we developed "formulaic-contrasts":
formulaic-contrasts.readthedocs.io/en/latest/
To compare sth, one needs to specify a contrast, e.g "on treatment vs baseline".
To make this easier, we developed "formulaic-contrasts":
formulaic-contrasts.readthedocs.io/en/latest/
Reposted by Gregor Sturm
📢 Preprint alert: #Benchmarking second-generation methods for cell-type #deconvolution of #transcriptomic data
t.co/MhLf5Eo3yY
A thread 🧵👇
1/
t.co/MhLf5Eo3yY
A thread 🧵👇
1/
https://doi.org/10.1101/2024.06.10.598226
t.co
November 12, 2024 at 1:41 PM
📢 Preprint alert: #Benchmarking second-generation methods for cell-type #deconvolution of #transcriptomic data
t.co/MhLf5Eo3yY
A thread 🧵👇
1/
t.co/MhLf5Eo3yY
A thread 🧵👇
1/
Reposted by Gregor Sturm
Explore the scverse Starter Pack!
Stay informed about the latest scverse events, software updates, and community news. Everything you need to know about foundational tools for single-cell omics analysis in one place.
go.bsky.app/UvFMa8d
Stay informed about the latest scverse events, software updates, and community news. Everything you need to know about foundational tools for single-cell omics analysis in one place.
go.bsky.app/UvFMa8d
November 16, 2024 at 1:59 PM
Explore the scverse Starter Pack!
Stay informed about the latest scverse events, software updates, and community news. Everything you need to know about foundational tools for single-cell omics analysis in one place.
go.bsky.app/UvFMa8d
Stay informed about the latest scverse events, software updates, and community news. Everything you need to know about foundational tools for single-cell omics analysis in one place.
go.bsky.app/UvFMa8d
Reposted by Gregor Sturm
And I made one for single cell which is needs more people, but is still a starting point 😅
go.bsky.app/U57Mar8
go.bsky.app/U57Mar8
November 11, 2024 at 7:29 AM
And I made one for single cell which is needs more people, but is still a starting point 😅
go.bsky.app/U57Mar8
go.bsky.app/U57Mar8

