
Gregor Sturm
@grst.bsky.social
Single Cell/Spatial. Cancer Immunology. Outdoor activities.
Core developer @scverse.bsky.social.
Working in Clinical Bioinformatics at Boehringer Ingelheim.
Formerly PhD student at Medical University of Innsbruck.
My private account.
github.com/grst
Core developer @scverse.bsky.social.
Working in Clinical Bioinformatics at Boehringer Ingelheim.
Formerly PhD student at Medical University of Innsbruck.
My private account.
github.com/grst
AFAIK, these differences are minor, numeric differences. I would consider them equivalent.
August 19, 2025 at 3:33 PM
AFAIK, these differences are minor, numeric differences. I would consider them equivalent.
Reposted by Gregor Sturm
Rogue Scholar
rogue-scholar.org
March 14, 2025 at 10:01 AM
Nice post!
How did you generate the doi-link for a blog post?
How did you generate the doi-link for a blog post?
March 14, 2025 at 9:52 AM
Nice post!
How did you generate the doi-link for a blog post?
How did you generate the doi-link for a blog post?
We try to avoid that by using this with preprocessed data only. All the heavy lifting is done with nextflow pipelines before. Datasets up to tens of GBs have worked well so far.
February 5, 2025 at 6:40 PM
We try to avoid that by using this with preprocessed data only. All the heavy lifting is done with nextflow pipelines before. Datasets up to tens of GBs have worked well so far.
Finally, many thanks to my colleagues @alexpeltzer.bsky.social, Daniel Schreyer and Tom Schwarzl for testing, adopting, and contributing to DSO.
February 5, 2025 at 6:32 PM
Finally, many thanks to my colleagues @alexpeltzer.bsky.social, Daniel Schreyer and Tom Schwarzl for testing, adopting, and contributing to DSO.
If you want to learn more, I'll be presenting this at a @nf-co.re bytesize talk: nf-co.re/events/2025/...

Bytesize: data science operations (DSO)
Gregor Sturm, Boehringer Ingelheim
nf-co.re
February 5, 2025 at 6:32 PM
If you want to learn more, I'll be presenting this at a @nf-co.re bytesize talk: nf-co.re/events/2025/...
We built this at @boehringerglobal.bsky.social to meet the quality standards required for biomarker analysis in clinical trials.
But I think this is useful for any kind of data analysis project.
But I think this is useful for any kind of data analysis project.
February 5, 2025 at 6:32 PM
We built this at @boehringerglobal.bsky.social to meet the quality standards required for biomarker analysis in clinical trials.
But I think this is useful for any kind of data analysis project.
But I think this is useful for any kind of data analysis project.
One of my favorite features: automated watermarking of all plots in a quarto report. Nobody gonna publish my plots anymore before I think they are ready.
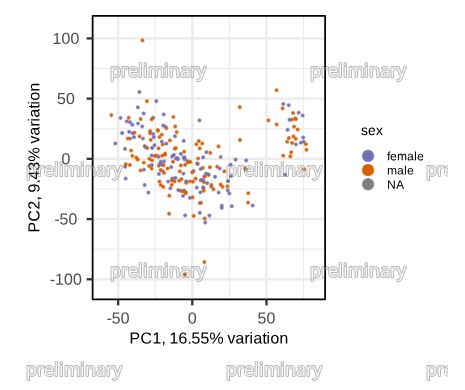
February 5, 2025 at 6:32 PM
One of my favorite features: automated watermarking of all plots in a quarto report. Nobody gonna publish my plots anymore before I think they are ready.
It brings together the best tools:
- git, for code versioning
- dvc, for data versioning and tracking inputs and outputs
- jinja2, for templates
- uv, for Python dep mgmt
- quarto, for authoring reports
- hiyapyco, for hierarchical YAML config
- pre-commit, for linting
- git, for code versioning
- dvc, for data versioning and tracking inputs and outputs
- jinja2, for templates
- uv, for Python dep mgmt
- quarto, for authoring reports
- hiyapyco, for hierarchical YAML config
- pre-commit, for linting
February 5, 2025 at 6:32 PM
It brings together the best tools:
- git, for code versioning
- dvc, for data versioning and tracking inputs and outputs
- jinja2, for templates
- uv, for Python dep mgmt
- quarto, for authoring reports
- hiyapyco, for hierarchical YAML config
- pre-commit, for linting
- git, for code versioning
- dvc, for data versioning and tracking inputs and outputs
- jinja2, for templates
- uv, for Python dep mgmt
- quarto, for authoring reports
- hiyapyco, for hierarchical YAML config
- pre-commit, for linting
(2) Finding the mistake, tracing it back to its origin, and fixing it was only possible because the data and scripts for building the atlas are publicly available and fully reproducible. github.com/icbi-lab/luca
GitHub - icbi-lab/luca: Single-cell Lung Cancer Atlas with 1.2M cells
Single-cell Lung Cancer Atlas with 1.2M cells. Contribute to icbi-lab/luca development by creating an account on GitHub.
github.com
January 19, 2025 at 10:54 AM
(2) Finding the mistake, tracing it back to its origin, and fixing it was only possible because the data and scripts for building the atlas are publicly available and fully reproducible. github.com/icbi-lab/luca
(1) Maintaining a data resource is very much like maintaining software. It is never "done" but constantly improving.
January 19, 2025 at 10:54 AM
(1) Maintaining a data resource is very much like maintaining software. It is never "done" but constantly improving.

