Mike Gallagher
@geneticsmike7.bsky.social
Geneticist and stem cell biologist studying epigenetics and neurological disease at Whitehead Institute and MIT.
Oops, here’s the right link www.biorxiv.org/content/10.1...

Systematic characterization of existing and novel inducible transgenic systems in human pluripotent stem cells after prolonged differentiation
The ability to control transgene expression both temporally and quantitatively in human-relevant cells and tissues is a cornerstone of biomedical research. Additionally, precise transgene control is c...
www.biorxiv.org
November 5, 2025 at 2:58 AM
Oops, here’s the right link www.biorxiv.org/content/10.1...
Great job market we scientists are in
November 5, 2025 at 1:37 AM
Great job market we scientists are in
20/ in summary, this "iPSC-Xon" system overcomes critical limitations in current transgene systems in hPSC-based work, and will benefit from additional improvements. I'm on the job market and would love to bring and improve this technology to other researchers!
November 2, 2025 at 2:35 PM
20/ in summary, this "iPSC-Xon" system overcomes critical limitations in current transgene systems in hPSC-based work, and will benefit from additional improvements. I'm on the job market and would love to bring and improve this technology to other researchers!
19/ strangely, leaky SALL1 is not due to unwanted splicing in the absence of drug, but rather the unspliced pre-mRNA somehow escapes the nucleus and is translated in-frame to make the full-length protein. We still don't know how this works.

November 2, 2025 at 2:35 PM
19/ strangely, leaky SALL1 is not due to unwanted splicing in the absence of drug, but rather the unspliced pre-mRNA somehow escapes the nucleus and is translated in-frame to make the full-length protein. We still don't know how this works.
18/ we thought larger transgenes (Cas9 is ~4kb) might be more prone to disrupting the Xon splicing switch, so we tried the ~4kb SALL1 TF, a critical driver of microglia homeostatic function. In contrast with EGFP, Xon-SALL1 is fully leaky and shows aberrant splicing patterns.
November 2, 2025 at 2:34 PM
18/ we thought larger transgenes (Cas9 is ~4kb) might be more prone to disrupting the Xon splicing switch, so we tried the ~4kb SALL1 TF, a critical driver of microglia homeostatic function. In contrast with EGFP, Xon-SALL1 is fully leaky and shows aberrant splicing patterns.
17/ we also show that we can get inducible gene editing with Xon hPSCs, but only if sgRNAs are introduced transiently (panels C-E). Cas9 must have enough leakiness for maximal editing with constitutive lentiviral sgRNA introduced into hPSCs (A-B)

November 2, 2025 at 2:32 PM
17/ we also show that we can get inducible gene editing with Xon hPSCs, but only if sgRNAs are introduced transiently (panels C-E). Cas9 must have enough leakiness for maximal editing with constitutive lentiviral sgRNA introduced into hPSCs (A-B)
16/ what about other genes? Here we show Xon works for inducible control of GRN expression, and we can overexpress GRN in iPSC-derived microglia, which already express boatloads of it. P2A peptides improve both GRN and SMAD, and importantly reduce N-terminal tag to a single aa.

November 2, 2025 at 2:31 PM
16/ what about other genes? Here we show Xon works for inducible control of GRN expression, and we can overexpress GRN in iPSC-derived microglia, which already express boatloads of it. P2A peptides improve both GRN and SMAD, and importantly reduce N-terminal tag to a single aa.
15/ a couple important points here: in addition to resisting silencing and being highly tunable, Xon adds only 24aa N-terminal tag (see below where we reduce it to 1aa) and needs only low nM drug doses, whereas degrons add ~500bp tags to BOTH termini to be effective, and need low uM drug doses.
November 2, 2025 at 2:30 PM
15/ a couple important points here: in addition to resisting silencing and being highly tunable, Xon adds only 24aa N-terminal tag (see below where we reduce it to 1aa) and needs only low nM drug doses, whereas degrons add ~500bp tags to BOTH termini to be effective, and need low uM drug doses.
14/ a note of caution is that while background EGFP is minor compared to even the lowest drug induced expression levels, this may need mitigation when using TFs or other strong cell state regulators

November 2, 2025 at 2:30 PM
14/ a note of caution is that while background EGFP is minor compared to even the lowest drug induced expression levels, this may need mitigation when using TFs or other strong cell state regulators
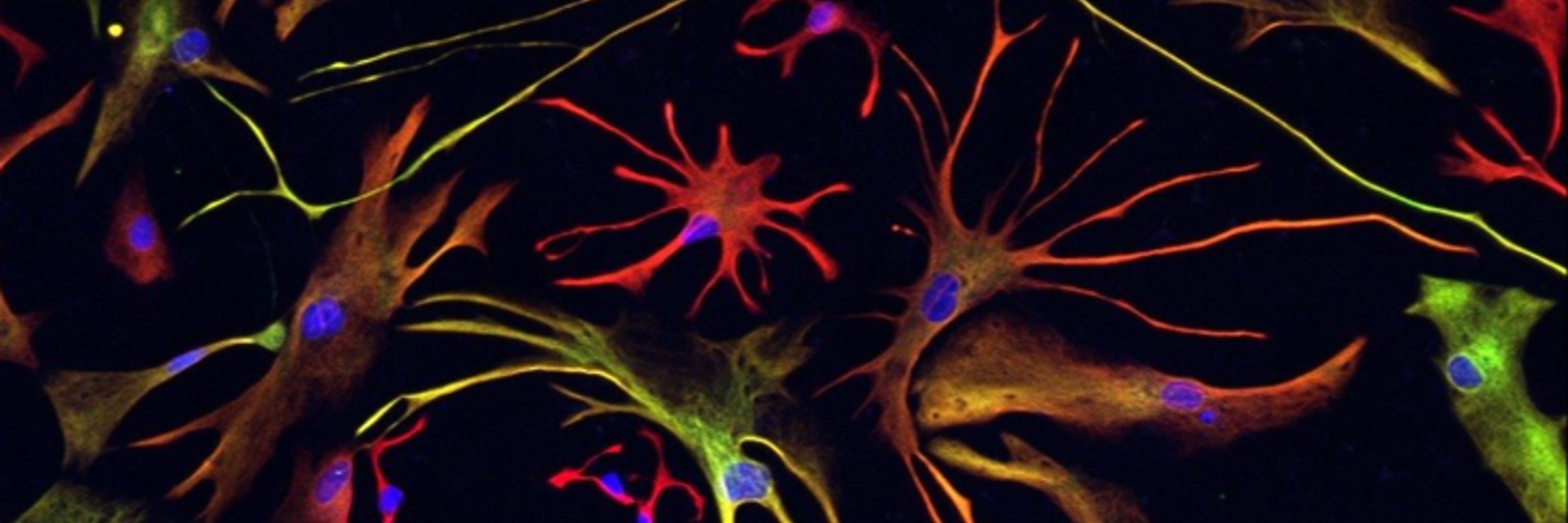
13/ Here's the commonly used H1 ESC background, differentiated into microglia with an embryoid body protocol (we used EB and monolayer)

November 2, 2025 at 2:29 PM
13/ Here's the commonly used H1 ESC background, differentiated into microglia with an embryoid body protocol (we used EB and monolayer)
12/ quantification shows how tunable this system really is! Below the 100nM dose, linear correlations between drug dose and EGFP is basically perfect both before and after differentiation

November 2, 2025 at 2:29 PM
12/ quantification shows how tunable this system really is! Below the 100nM dose, linear correlations between drug dose and EGFP is basically perfect both before and after differentiation
11/ again with AAVS1-targeted EGFP, what we saw was remarkable: first time we've ever seen robust inducibility after long differentiations, this time 4-7 weeks, across hPSC backgrounds and differentiation protocols!

November 2, 2025 at 2:28 PM
11/ again with AAVS1-targeted EGFP, what we saw was remarkable: first time we've ever seen robust inducibility after long differentiations, this time 4-7 weeks, across hPSC backgrounds and differentiation protocols!
10/ we then adapted the recently described "Xon" splicing switch, from this paper: pmc.ncbi.nlm.nih.gov/articles/PMC...
Could this approach work in hPSCs, resist silencing and allow for robust inducibility in differentiated microglia and macrophages?
Could this approach work in hPSCs, resist silencing and allow for robust inducibility in differentiated microglia and macrophages?

Regulated control of gene therapies by drug-induced splicing
So far, gene therapies have relied on complex constructs that cannot be finely controlled1,2. Here we report a universal switch element that enables precise control of gene replacement or gene editing after exposure to a small molecule. The ...
pmc.ncbi.nlm.nih.gov
November 2, 2025 at 2:27 PM
10/ we then adapted the recently described "Xon" splicing switch, from this paper: pmc.ncbi.nlm.nih.gov/articles/PMC...
Could this approach work in hPSCs, resist silencing and allow for robust inducibility in differentiated microglia and macrophages?
Could this approach work in hPSCs, resist silencing and allow for robust inducibility in differentiated microglia and macrophages?
9/ we also looked at leakiness, and leaky EGFP varied by system, with Tet-On being pretty tightly controlled (although silenced later on), and degrons showing variability. Note lack of degron tunability in panel C.

November 2, 2025 at 2:27 PM
9/ we also looked at leakiness, and leaky EGFP varied by system, with Tet-On being pretty tightly controlled (although silenced later on), and degrons showing variability. Note lack of degron tunability in panel C.
8b/ as you can see above, degrons vary in performance, but all show no tunability and only modest inducibility after differentiation.
November 2, 2025 at 2:26 PM
8b/ as you can see above, degrons vary in performance, but all show no tunability and only modest inducibility after differentiation.
8/ We next tested degrons, as this have been used successfully for transgene expression specifically in fully differentiated cells. However, their leakiness, tunability, and inducibility hasn't been thoroughly tested.

November 2, 2025 at 2:24 PM
8/ We next tested degrons, as this have been used successfully for transgene expression specifically in fully differentiated cells. However, their leakiness, tunability, and inducibility hasn't been thoroughly tested.
7/ all approaches used EGFP after AAVS1 safe harbor targeting. While these systems worked in hPSCs (albeit with slower kinetics), they still got silenced during differentiation

November 2, 2025 at 2:24 PM
7/ all approaches used EGFP after AAVS1 safe harbor targeting. While these systems worked in hPSCs (albeit with slower kinetics), they still got silenced during differentiation
6/ we then tried to fix this by adding a Tet-Off-TET1 fusion protein, as DNA methylation has been proposed to underlie Tet-On silencing. Here's the idea, with existing systems (top) getting silenced before DOX is added, and new systems (bottom) we hoped would fix it

November 2, 2025 at 2:23 PM
6/ we then tried to fix this by adding a Tet-Off-TET1 fusion protein, as DNA methylation has been proposed to underlie Tet-On silencing. Here's the idea, with existing systems (top) getting silenced before DOX is added, and new systems (bottom) we hoped would fix it
5/ First we showed that Tet-On-based transgenes are silenced early during microglia or macrophage differentiation, regardless of transgene delivery method, transgene components, hPSC background, or differentiation protocol

November 2, 2025 at 2:21 PM
5/ First we showed that Tet-On-based transgenes are silenced early during microglia or macrophage differentiation, regardless of transgene delivery method, transgene components, hPSC background, or differentiation protocol
4/ We compared multiple existing systems, tried to fix the ones that don't work, and developed a new system that overcomes many existing problems, using #microglia and #macrophages as test cases #Immunotherapy #neuroinflammation LINK:https://www.biorxiv.org/content/10.1101/2025.10.17.683097v1
November 2, 2025 at 2:21 PM
4/ We compared multiple existing systems, tried to fix the ones that don't work, and developed a new system that overcomes many existing problems, using #microglia and #macrophages as test cases #Immunotherapy #neuroinflammation LINK:https://www.biorxiv.org/content/10.1101/2025.10.17.683097v1
3/ this is a major problem for both basic biomedical research in disease-relevant cells/tissues, as well as CRISPR screens and other systematic gene/pathway dissections that must be done with careful control at the right stage of differentiation. #bioengineering #genomics #stemcells
November 2, 2025 at 2:20 PM
3/ this is a major problem for both basic biomedical research in disease-relevant cells/tissues, as well as CRISPR screens and other systematic gene/pathway dissections that must be done with careful control at the right stage of differentiation. #bioengineering #genomics #stemcells
2/ we cannot currently alter genes and pathways in many differentiated hPSC-derived cell types without potential effects on the differentiation itself, due to transgene silencing, lack of transgene tunability, and toxicity or unwanted effects of electroporation and transduction #ESC #iPSC #synbio
November 2, 2025 at 2:19 PM
Is it the same story for Science Translational Medicine and Science Immunology?
October 27, 2025 at 3:49 PM
Is it the same story for Science Translational Medicine and Science Immunology?
Then traditional Google Scholar should be fine?
October 26, 2025 at 1:27 PM
Then traditional Google Scholar should be fine?

