Any lab using @flybase.bsky.social please donate using the link in post below.
This incredible community, on whose backs our #Drosophila labs depend, can't be left out to dry.
www.philanthropy.cam.ac.uk/give-to-camb...
If you are excited about combining AI and machine learning with fundamental biology, we would love to hear from you!
https://vib.ai/en/opportunities#/job-description/110906

If you are excited about combining AI and machine learning with fundamental biology, we would love to hear from you!
https://vib.ai/en/opportunities#/job-description/110906
If you are excited about combining AI and machine learning with fundamental biology, we would love to hear from you!
https://vib.ai/en/opportunities#/job-description/110906

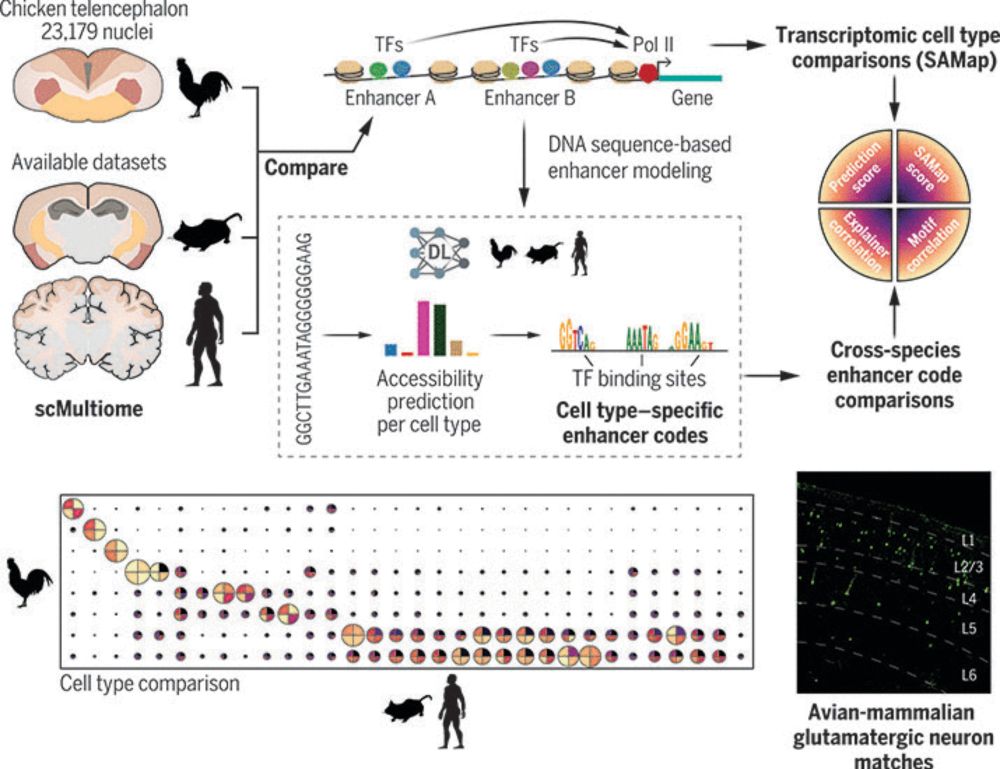
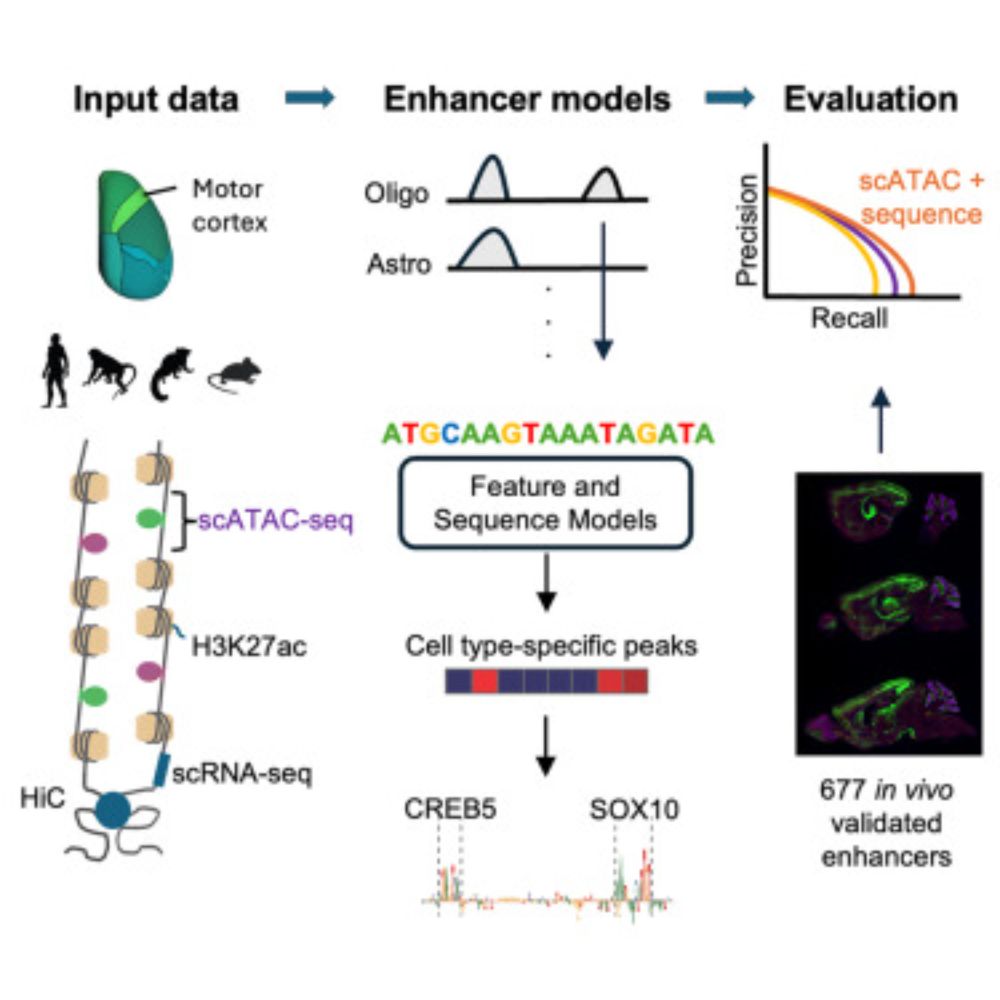
1) CREsted: to train sequence-to-function deep learning models on scATAC-seq atlases, and use them to decipher enhancer logic and design synthetic enhancers. This has been a wonderful lab-wide collaborative effort. www.biorxiv.org/content/10.1...

1) CREsted: to train sequence-to-function deep learning models on scATAC-seq atlases, and use them to decipher enhancer logic and design synthetic enhancers. This has been a wonderful lab-wide collaborative effort. www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
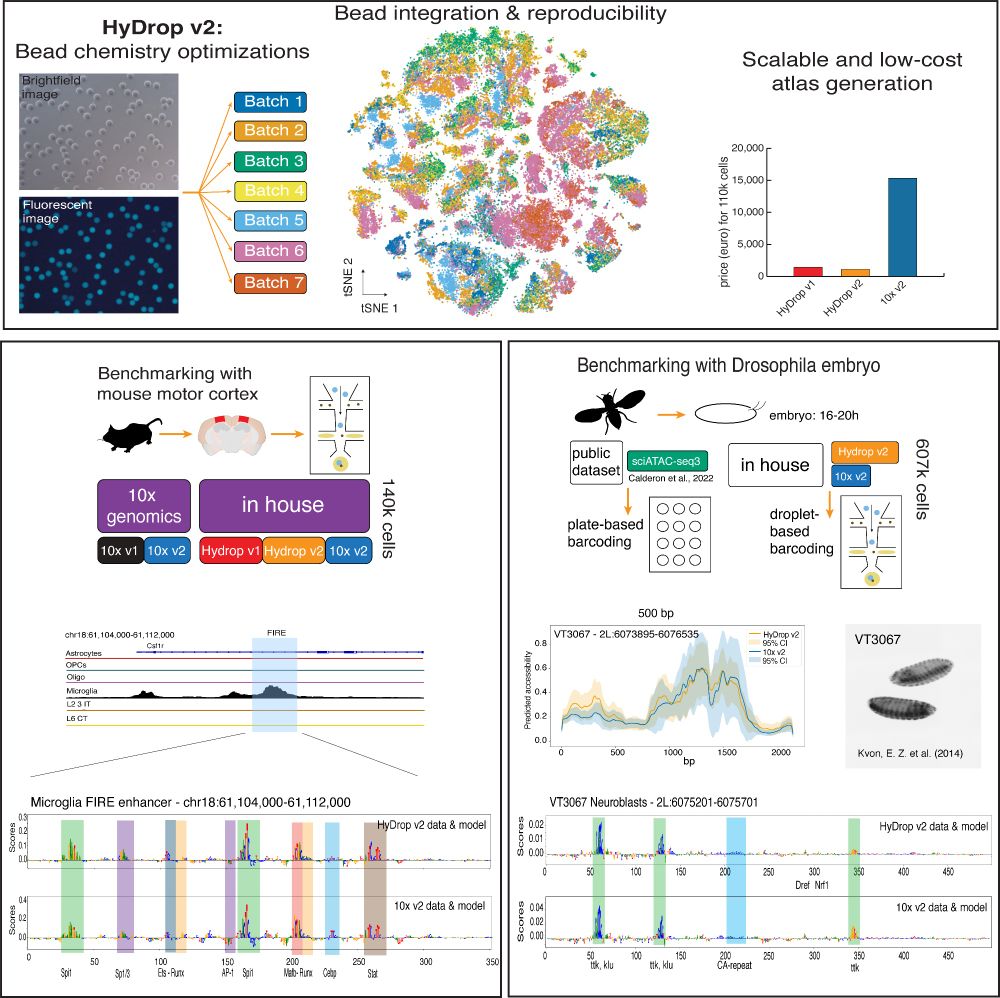
www.biorxiv.org/content/10.1...



From conventional machine learning methods to CNNs and using models as oracles/generative AI for synthetic enhancer design!
@natrevbioeng.bsky.social
www.nature.com/articles/s44...
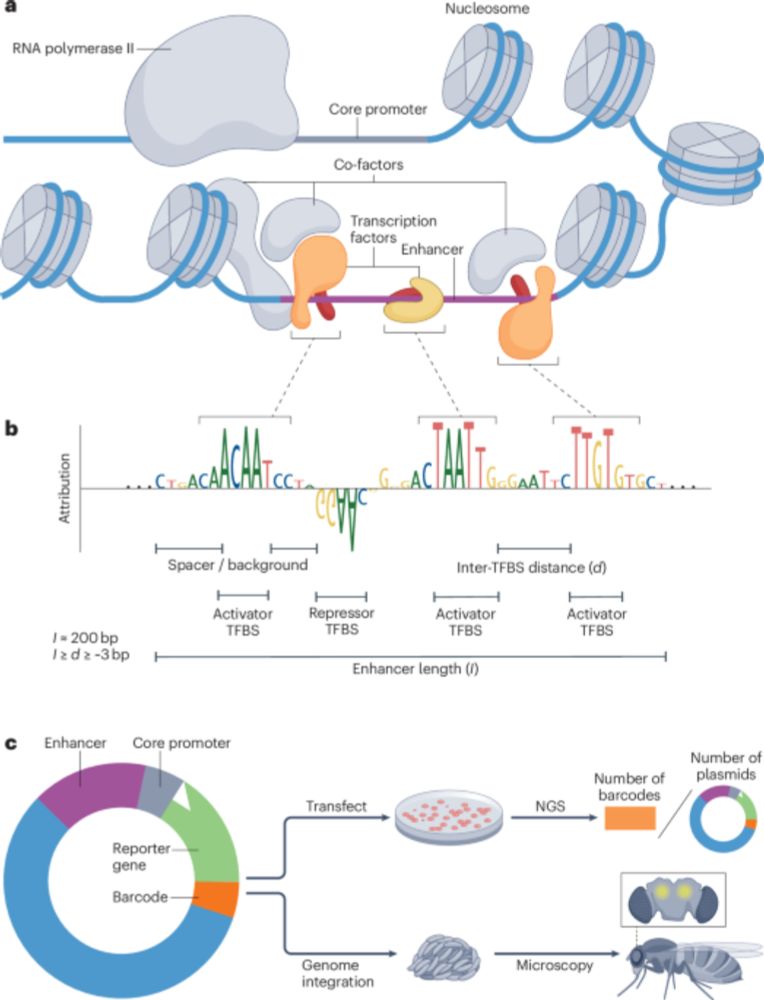
From conventional machine learning methods to CNNs and using models as oracles/generative AI for synthetic enhancer design!
@natrevbioeng.bsky.social
www.nature.com/articles/s44...
www.science.org/doi/10.1126/...
www.science.org/doi/10.1126/...