
To learn more about FROs, visit convergentresearch.org.
huggingface.co/blog/hugging...

huggingface.co/blog/hugging...
evebio.org/pharmome-dat...
Here's what's new: 🧵

evebio.org/pharmome-dat...
Here's what's new: 🧵
Learn more: evebio.org/pharmome-dat...

Learn more: evebio.org/pharmome-dat...
www.youtube.com/watch?v=WUU1...

www.youtube.com/watch?v=WUU1...
prism.cultivarium.org/protocols/20...
prism.cultivarium.org/protocols/20...
prism.cultivarium.org/protocols/20...
prism.cultivarium.org/protocols/20...
prism.cultivarium.org/protocols/20...
prism.cultivarium.org/protocols/20...
Plasmids (also on GitHub):
www.addgene.org/199102/
www.addgene.org/89477/
Genome (also on GitHub):
... stuck in submission at NCBI... for now.
If you're interested in using the mutant library yourself, reach out!

Plasmids (also on GitHub):
www.addgene.org/199102/
www.addgene.org/89477/
Genome (also on GitHub):
... stuck in submission at NCBI... for now.
If you're interested in using the mutant library yourself, reach out!
We identified a plasmid vector for the strain and generated a large RB-TnSeq library, screening for genes impacting plastic degradation.
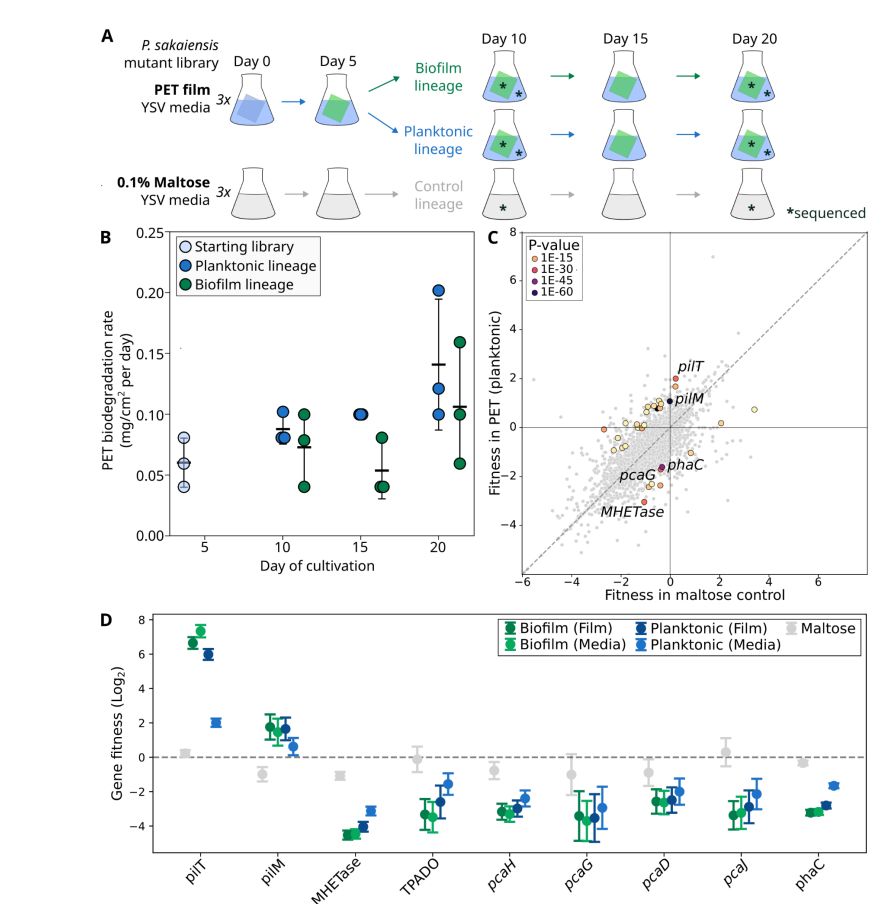
We identified a plasmid vector for the strain and generated a large RB-TnSeq library, screening for genes impacting plastic degradation.
Had fun writing this, ping me if interested in other search engines. #TeamMassSpec
www.parallelsq.org/blog/fantast...

Had fun writing this, ping me if interested in other search engines. #TeamMassSpec
www.parallelsq.org/blog/fantast...

prism.cultivarium.org/protocols/20...
We got to cite the E = mc² paper!
Kidding :)
The important part is that combinations of isotopes unlock 100-1000x gains in protein measurement speed.
Here's how: www.biorxiv.org/content/10.1...
#TeamMassSpec #Proteomics 🧪

We got to cite the E = mc² paper!
Kidding :)
The important part is that combinations of isotopes unlock 100-1000x gains in protein measurement speed.
Here's how: www.biorxiv.org/content/10.1...
#TeamMassSpec #Proteomics 🧪
We got to cite the E = mc² paper!
Kidding :)
The important part is that combinations of isotopes unlock 100-1000x gains in protein measurement speed.
Here's how: www.biorxiv.org/content/10.1...
#TeamMassSpec #Proteomics 🧪


/n
www.nature.com/articles/d41...

/n
www.nature.com/articles/d41...
- Simply not feasible with linearly scaling approaches and the resources we have.
timePlex makes it feasible, and I cannot stop thinking about it.
- Simply not feasible with linearly scaling approaches and the resources we have.
timePlex makes it feasible, and I cannot stop thinking about it.

Our team, led by Jason Derks, enabled combinatorial-scaling of mass spec proteomics throughput by developing multiplexing in the time domain, timePlex.
1/n

Our team, led by Jason Derks, enabled combinatorial-scaling of mass spec proteomics throughput by developing multiplexing in the time domain, timePlex.
1/n
Use the 𝚜𝚒𝚖𝚙? "Try this" suggestion to insert the precise 𝚜𝚒𝚖𝚙 𝚘𝚗𝚕𝚢 call into your proof.
Learn more: lean-lang.org/theorem_prov...
#LeanLang #LeanProver #ProofAssistant

Use the 𝚜𝚒𝚖𝚙? "Try this" suggestion to insert the precise 𝚜𝚒𝚖𝚙 𝚘𝚗𝚕𝚢 call into your proof.
Learn more: lean-lang.org/theorem_prov...
#LeanLang #LeanProver #ProofAssistant
www.youtube.com/watch?v=2rLB...

www.youtube.com/watch?v=2rLB...
Read the book here: lean-lang.org/theorem_prov...
#LeanLang #LeanProver #Mathematics

Read the book here: lean-lang.org/theorem_prov...
#LeanLang #LeanProver #Mathematics

