
Gautam Shirsekar
@coevolution.bsky.social
Plant pathologist | Genomics | Coevolution | Wild plant pathosystems | Weigelworld, Max Planck Institute for Biology | Asst. Prof. at the University of Tennessee, Knoxville (EPP)| www.coevolutionlab.org
Pinned
Gautam Shirsekar
@coevolution.bsky.social
· Aug 14
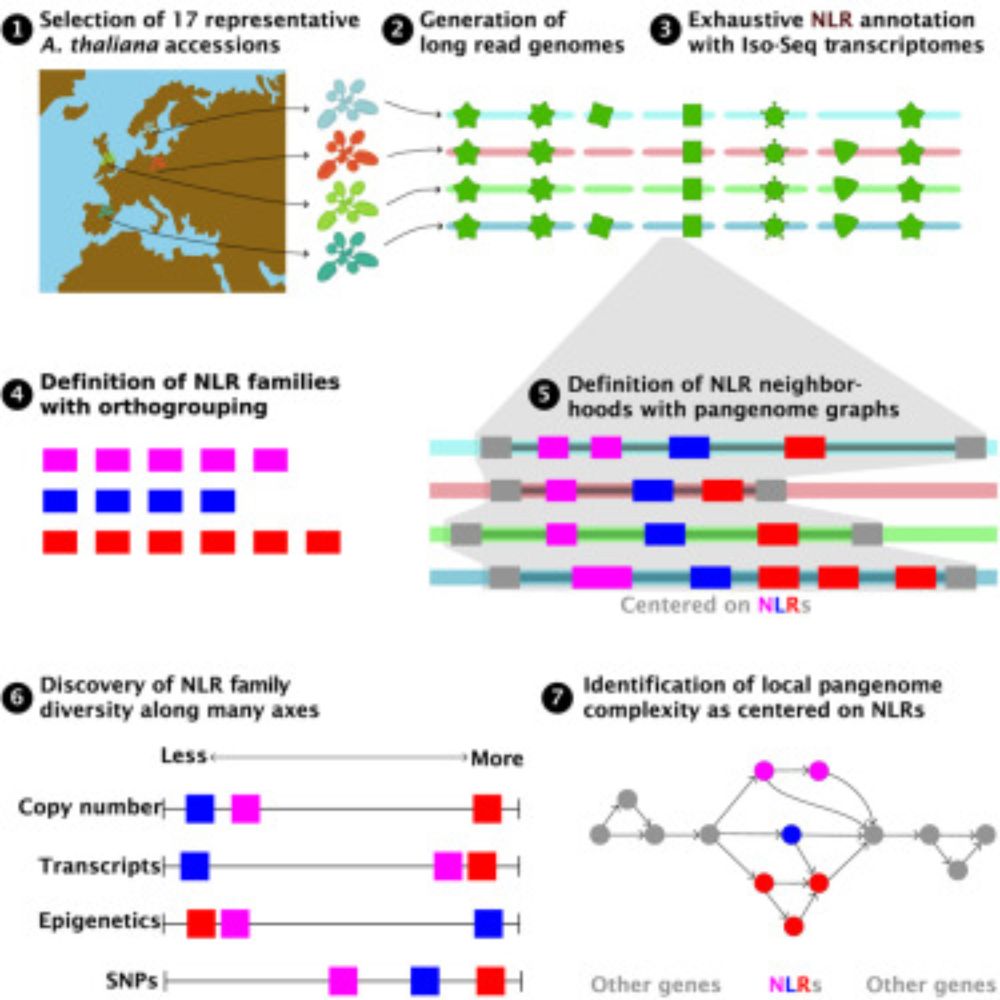
~ 5 and 1/2 years ago, my then postdoc advisor @plantevolution.bsky.social gave me complete freedom to assemble a team and interrogate NLR diversity in pangenomic context. We used @pacbio.bsky.social CCS and Iso-Seq, to annotate genomes with pathogen-challenged transcriptome. We then manually.....
Reposted by Gautam Shirsekar
New Correspondence: "Rethinking the need for field trials" rdcu.be/eM96P

October 28, 2025 at 7:12 PM
New Correspondence: "Rethinking the need for field trials" rdcu.be/eM96P
Reposted by Gautam Shirsekar
2/2 Our lab pioneered the use of genome graphs already 16 years ago! Read this oldie-but-goldie from @labschneeberger.bsky.social and Jörg Hagmann: Simultaneous alignment of short reads against multiple genomes
genomebiology.biomedcentral.com/articles/10....
genomebiology.biomedcentral.com/articles/10....

October 27, 2025 at 5:53 PM
2/2 Our lab pioneered the use of genome graphs already 16 years ago! Read this oldie-but-goldie from @labschneeberger.bsky.social and Jörg Hagmann: Simultaneous alignment of short reads against multiple genomes
genomebiology.biomedcentral.com/articles/10....
genomebiology.biomedcentral.com/articles/10....
Reposted by Gautam Shirsekar
1/2 Want to become up to date with pangenomes and genome graphs and their history? Check out this fantastic review by @zbao.bsky.social!
Complexity welcome: Pangenome graphs for comprehensive population genomics
#pangenomes #plantscience #genomegraphs
www.cambridge.org/core/journal...
Complexity welcome: Pangenome graphs for comprehensive population genomics
#pangenomes #plantscience #genomegraphs
www.cambridge.org/core/journal...

October 27, 2025 at 5:53 PM
1/2 Want to become up to date with pangenomes and genome graphs and their history? Check out this fantastic review by @zbao.bsky.social!
Complexity welcome: Pangenome graphs for comprehensive population genomics
#pangenomes #plantscience #genomegraphs
www.cambridge.org/core/journal...
Complexity welcome: Pangenome graphs for comprehensive population genomics
#pangenomes #plantscience #genomegraphs
www.cambridge.org/core/journal...
@annaliisalaine.bsky.social
Does this look familiar to you? From Cherokee National Forest
Does this look familiar to you? From Cherokee National Forest


September 16, 2025 at 3:58 PM
@annaliisalaine.bsky.social
Does this look familiar to you? From Cherokee National Forest
Does this look familiar to you? From Cherokee National Forest
Reposted by Gautam Shirsekar
Out after peer review, collaborative study from Nordborg & Weigel labs with help from many others. Not the largest collection of new Arabidopsis thaliana genomes, but we hopefully put forward some good ideas for how to think about pangenomes and their analysis!
www.nature.com/articles/s41...
www.nature.com/articles/s41...

August 20, 2025 at 6:23 AM
Out after peer review, collaborative study from Nordborg & Weigel labs with help from many others. Not the largest collection of new Arabidopsis thaliana genomes, but we hopefully put forward some good ideas for how to think about pangenomes and their analysis!
www.nature.com/articles/s41...
www.nature.com/articles/s41...
Reposted by Gautam Shirsekar
Plant NLR evolution
Annotated, divergent A. thaliana genomes &pangenome graph approaches describe genomic neighborhoods of NLRs, revealing evolutionary footprints in form of “diversity in diversity” at these loci
Weigel @mpi-bio-fml.bsky.social @coevolution.bsky.social
www.cell.com/cell-host-mi...
Annotated, divergent A. thaliana genomes &pangenome graph approaches describe genomic neighborhoods of NLRs, revealing evolutionary footprints in form of “diversity in diversity” at these loci
Weigel @mpi-bio-fml.bsky.social @coevolution.bsky.social
www.cell.com/cell-host-mi...
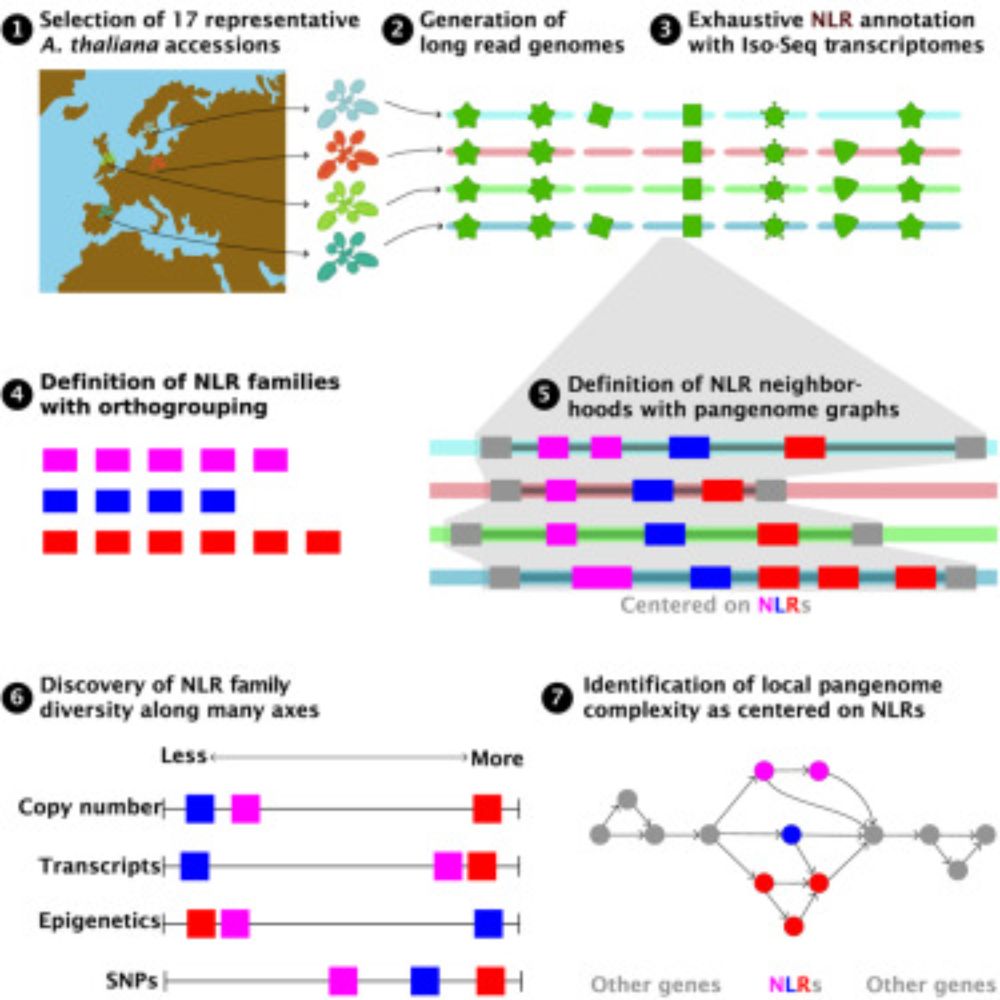
Pangenomic context reveals the extent of intraspecific plant NLR evolution
Individual- and population-level diversity is required for pathogen defense by nucleotide-binding
site leucine-rich repeat (NLR) proteins. Teasdale et al. leverage annotated, divergent
A. thaliana gen...
www.cell.com
August 15, 2025 at 3:08 PM
Plant NLR evolution
Annotated, divergent A. thaliana genomes &pangenome graph approaches describe genomic neighborhoods of NLRs, revealing evolutionary footprints in form of “diversity in diversity” at these loci
Weigel @mpi-bio-fml.bsky.social @coevolution.bsky.social
www.cell.com/cell-host-mi...
Annotated, divergent A. thaliana genomes &pangenome graph approaches describe genomic neighborhoods of NLRs, revealing evolutionary footprints in form of “diversity in diversity” at these loci
Weigel @mpi-bio-fml.bsky.social @coevolution.bsky.social
www.cell.com/cell-host-mi...
~ 5 and 1/2 years ago, my then postdoc advisor @plantevolution.bsky.social gave me complete freedom to assemble a team and interrogate NLR diversity in pangenomic context. We used @pacbio.bsky.social CCS and Iso-Seq, to annotate genomes with pathogen-challenged transcriptome. We then manually.....
August 14, 2025 at 9:13 PM
~ 5 and 1/2 years ago, my then postdoc advisor @plantevolution.bsky.social gave me complete freedom to assemble a team and interrogate NLR diversity in pangenomic context. We used @pacbio.bsky.social CCS and Iso-Seq, to annotate genomes with pathogen-challenged transcriptome. We then manually.....
Reposted by Gautam Shirsekar
Out after peer review now, follow up from our 2019 pna-NLRome paper (which was based on enrichment and long-read sequencing). It is remarkable how much more can be learned with complete genome sequences. Next. pan-NLRome from hundreds of A. thaliana long-read genomes! Big thanks to the entire team!
Happy to be able to finally share our NLR pangenome paper, out now in CHM.
"Pangenomic context reveals the extent of intraspecific plant NLR evolution"
www.cell.com/cell-host-mi...
#plantscience #plantimmunity #pangenomes #science #nlr
"Pangenomic context reveals the extent of intraspecific plant NLR evolution"
www.cell.com/cell-host-mi...
#plantscience #plantimmunity #pangenomes #science #nlr
Pangenomic context reveals the extent of intraspecific plant NLR evolution
Individual- and population-level diversity is required for pathogen defense by nucleotide-binding
site leucine-rich repeat (NLR) proteins. Teasdale et al. leverage annotated, divergent
A. thaliana gen...
www.cell.com
August 14, 2025 at 1:42 PM
Out after peer review now, follow up from our 2019 pna-NLRome paper (which was based on enrichment and long-read sequencing). It is remarkable how much more can be learned with complete genome sequences. Next. pan-NLRome from hundreds of A. thaliana long-read genomes! Big thanks to the entire team!
Reposted by Gautam Shirsekar
1/2 What's best: a field-first or lab-first approach? No easy answers but differences between lab and field should not be seen as failure but motivate further inquiry and allow complementary discovery. Read our thoughts on this here:
www.sciencedirect.com/science/arti...
www.sciencedirect.com/science/arti...

Lab to field: Challenges and opportunities for plant biology
Plant-microbe research offers many choices of model and strain and whether a field-first or lab-first approach is best. However, differences between l…
www.sciencedirect.com
August 14, 2025 at 3:44 PM
1/2 What's best: a field-first or lab-first approach? No easy answers but differences between lab and field should not be seen as failure but motivate further inquiry and allow complementary discovery. Read our thoughts on this here:
www.sciencedirect.com/science/arti...
www.sciencedirect.com/science/arti...
Reposted by Gautam Shirsekar
Feel free to ask any q's here
August 14, 2025 at 5:55 AM
Feel free to ask any q's here
Reposted by Gautam Shirsekar
We show that pangenome graphs provide a flexible way to define the context within which NLR evolution occurs, and that graph metrics capture diversity in both sequence and structure, and show that such diversity is centered on NLRs themselves

August 14, 2025 at 5:55 AM
We show that pangenome graphs provide a flexible way to define the context within which NLR evolution occurs, and that graph metrics capture diversity in both sequence and structure, and show that such diversity is centered on NLRs themselves
Reposted by Gautam Shirsekar
Long story short: we assemble 17 representative Arabidopsis thaliana genomes, exhaustively annotate NLRs with long read evidence & manual curation, and assay many measures of NLR diversity.

August 14, 2025 at 5:55 AM
Long story short: we assemble 17 representative Arabidopsis thaliana genomes, exhaustively annotate NLRs with long read evidence & manual curation, and assay many measures of NLR diversity.
'Hoods are out!!! Big thank you to @plantevolution.bsky.social for the support throughout. Luisa, @kdm9.bsky.social , @aconga.bsky.social , @hajkdrost.bsky.social it was intellectually stimulating ride with you, so congratulations!!!! #diversity #NLR #immune #pan-genome #graphs #networktheory
Happy to be able to finally share our NLR pangenome paper, out now in CHM.
"Pangenomic context reveals the extent of intraspecific plant NLR evolution"
www.cell.com/cell-host-mi...
#plantscience #plantimmunity #pangenomes #science #nlr
"Pangenomic context reveals the extent of intraspecific plant NLR evolution"
www.cell.com/cell-host-mi...
#plantscience #plantimmunity #pangenomes #science #nlr

Pangenomic context reveals the extent of intraspecific plant NLR evolution
Individual- and population-level diversity is required for pathogen defense by nucleotide-binding
site leucine-rich repeat (NLR) proteins. Teasdale et al. leverage annotated, divergent
A. thaliana gen...
www.cell.com
August 14, 2025 at 8:22 PM
'Hoods are out!!! Big thank you to @plantevolution.bsky.social for the support throughout. Luisa, @kdm9.bsky.social , @aconga.bsky.social , @hajkdrost.bsky.social it was intellectually stimulating ride with you, so congratulations!!!! #diversity #NLR #immune #pan-genome #graphs #networktheory
Come join EEB at the University of Tennessee at Knoxville.
Evolutionary Genomics, Assistant Professor, Ecology & Evolutionary Biology, Fall 2026
apply.interfolio.com/170735
functional and evolutionary genomics in any system 🌱, 🦠, or 🦌 (including 👫)
Evolutionary Genomics, Assistant Professor, Ecology & Evolutionary Biology, Fall 2026
apply.interfolio.com/170735
functional and evolutionary genomics in any system 🌱, 🦠, or 🦌 (including 👫)
Apply - Interfolio
{{$ctrl.$state.data.pageTitle}} - Apply - Interfolio
apply.interfolio.com
July 28, 2025 at 5:46 PM
Come join EEB at the University of Tennessee at Knoxville.
Evolutionary Genomics, Assistant Professor, Ecology & Evolutionary Biology, Fall 2026
apply.interfolio.com/170735
functional and evolutionary genomics in any system 🌱, 🦠, or 🦌 (including 👫)
Evolutionary Genomics, Assistant Professor, Ecology & Evolutionary Biology, Fall 2026
apply.interfolio.com/170735
functional and evolutionary genomics in any system 🌱, 🦠, or 🦌 (including 👫)

May 12, 2025 at 10:08 PM
In the Smokies, on a wild grape hunt




May 5, 2025 at 1:04 AM
In the Smokies, on a wild grape hunt
1/6 Today is Labor Day in USA and we decided to release the results of our labor and rigor on NLR evolution as a preprint. www.biorxiv.org/content/10.1...

Pangenomic context reveals the extent of intraspecific plant NLR evolution
bioRxiv - the preprint server for biology, operated by Cold Spring Harbor Laboratory, a research and educational institution
www.biorxiv.org
September 2, 2024 at 10:08 PM
1/6 Today is Labor Day in USA and we decided to release the results of our labor and rigor on NLR evolution as a preprint. www.biorxiv.org/content/10.1...
Wild Flower Festival, Frozen Head State Park, #TN #statepark
#TNPS, Tennessee Native Plant Society #naturalist
#TNPS, Tennessee Native Plant Society #naturalist




April 15, 2024 at 12:48 AM
Wild Flower Festival, Frozen Head State Park, #TN #statepark
#TNPS, Tennessee Native Plant Society #naturalist
#TNPS, Tennessee Native Plant Society #naturalist
Wild Flower Festival, Frozen Head State Park, #TN #statepark
#TNPS, Tennessee Native Plant Society #naturalist
#TNPS, Tennessee Native Plant Society #naturalist




April 15, 2024 at 12:47 AM
Wild Flower Festival, Frozen Head State Park, #TN #statepark
#TNPS, Tennessee Native Plant Society #naturalist
#TNPS, Tennessee Native Plant Society #naturalist
Wild Flower Festival, Frozen Head State Park, #TN #statepark
#TNPS, Tennessee Native Plant Society #naturalist
#TNPS, Tennessee Native Plant Society #naturalist




April 15, 2024 at 12:46 AM
Wild Flower Festival, Frozen Head State Park, #TN #statepark
#TNPS, Tennessee Native Plant Society #naturalist
#TNPS, Tennessee Native Plant Society #naturalist



