“A scalable framework to link rare human variants to disease phenotypes using pooled prime editing”
Friday Oct 17th at 1:40pm, Rm205ABC
You can also catch up with Chloé at her poster, 5002T, Thursday 2:30-4:30
“A scalable framework to link rare human variants to disease phenotypes using pooled prime editing”
Friday Oct 17th at 1:40pm, Rm205ABC
You can also catch up with Chloé at her poster, 5002T, Thursday 2:30-4:30
Check out our talks over the next few days - all unpublished stories.
Kicking things off is @michaelherger.bsky.social presenting "Saturation mutagenesis of 37 human splicing factor genes with pooled prime editing". Today @2pm, Rm205abc
Also 👇
Check out our talks over the next few days - all unpublished stories.
Kicking things off is @michaelherger.bsky.social presenting "Saturation mutagenesis of 37 human splicing factor genes with pooled prime editing". Today @2pm, Rm205abc
Also 👇

www.crick.ac.uk/careers-and-...

www.crick.ac.uk/careers-and-...
Feel free to reach out if you’d like to hear more about my experience in the group!
We are pleased to be recruiting this year. 👇
www.crick.ac.uk/careers-stud...
Feel free to reach out if you’d like to hear more about my experience in the group!
crick.wd3.myworkdayjobs.com/External/job...
Please apply if you have a background in functional genomics or a related field and are eager to develop methods to map variant effects at scale.
Big kudos to my co-organisers for making it all possible 🌟
👉 Come and join our amazing community: londonomics.co.uk
Big kudos to my co-organisers for making it all possible 🌟
👉 Come and join our amazing community: londonomics.co.uk
Read more about it👇
www.crick.ac.uk/news/2025-09...

Read more about it👇
If you are interested in the development of high-throughput assays to test non-coding variant effects come join us at Session 53 on Thursday, October 16th!

If you are interested in the development of high-throughput assays to test non-coding variant effects come join us at Session 53 on Thursday, October 16th!
Meet the panellists for one of the upcoming live episode recordings of our podcast, A Question of Science:
@profjoyceharper.bsky.social, @lucyvandewiel.bsky.social,
@nmoris.bsky.social and @gunes-taylor.bsky.social.
🎟️ Book now ⬇️
🔗 lostintv.com/tv-show?id=1...

Meet the panellists for one of the upcoming live episode recordings of our podcast, A Question of Science:
@profjoyceharper.bsky.social, @lucyvandewiel.bsky.social,
@nmoris.bsky.social and @gunes-taylor.bsky.social.
🎟️ Book now ⬇️
🔗 lostintv.com/tv-show?id=1...
tinyurl.com/3j9r56s8
@rociorius.bsky.social @yuyangchen.bsky.social @gregfindlay.bsky.social @dgmacarthur.bsky.social @cassimons.bsky.social @nickywhiffin.bsky.social

Thanks to the lab, Nicole, @lcubes.bsky.social @chloeterwagne.bsky.social and Megan), our great collaborators, and @crick.ac.uk & @cancerresearchuk.org for vital funding. 🙏 END/16

Thanks to the lab, Nicole, @lcubes.bsky.social @chloeterwagne.bsky.social and Megan), our great collaborators, and @crick.ac.uk & @cancerresearchuk.org for vital funding. 🙏 END/16
Saturation genome editing of BRCA1 across cell types, packed with insights 👇
www.medrxiv.org/content/10.1...
Saturation genome editing of BRCA1 across cell types accurately resolves cancer risk.
Led by the amazing Phoebe Dace. This one’s packed full of data, so check out the paper. Quick highlights… 🧵 1/n

Saturation genome editing of BRCA1 across cell types, packed with insights 👇
➡️Christina Kajba @ckajba.bsky.social & Michael Herger @michaelherger.bsky.social — pooled prime editing screens in haploid human cells
➡️Jon Acosta — multiplexed in vivo base editing screens
#GeneticVariants #PrimeEditing
ℹ️https://www.varianteffect.org/seminar-series/

➡️Christina Kajba @ckajba.bsky.social & Michael Herger @michaelherger.bsky.social — pooled prime editing screens in haploid human cells
➡️Jon Acosta — multiplexed in vivo base editing screens
#GeneticVariants #PrimeEditing
ℹ️https://www.varianteffect.org/seminar-series/
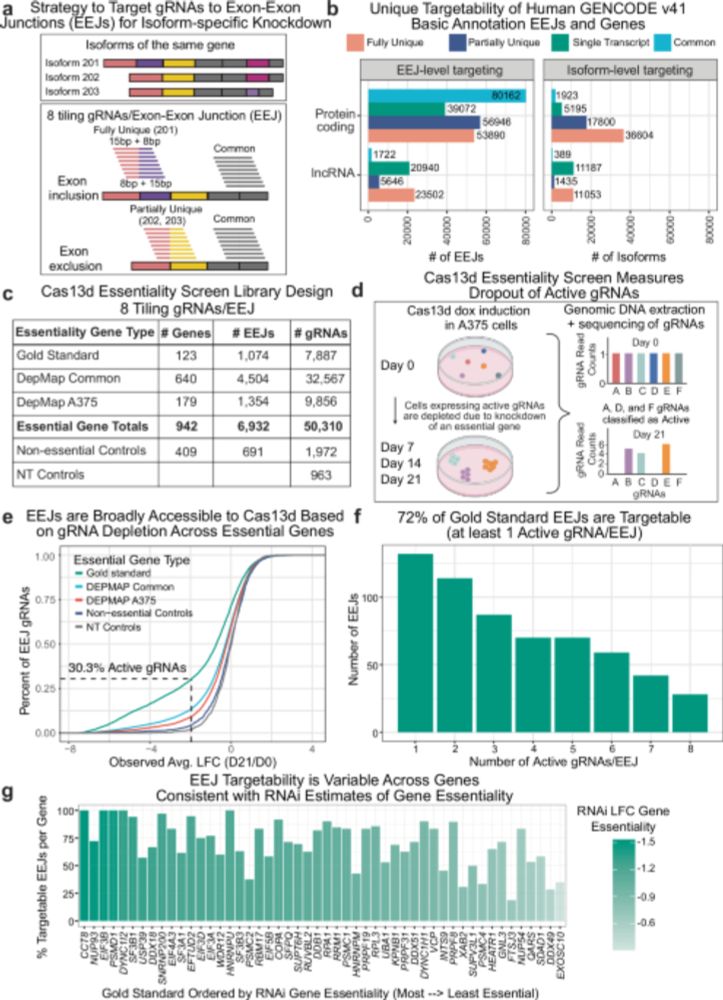
🧬 Join us: londonomics.co.uk/index.php/ev...
Abstract deadline: August 12
👩💻 Don’t miss your chance to present your work and connect with the community!
See you there!
🧬 Join us: londonomics.co.uk/index.php/ev...
Abstract deadline: August 12
👩💻 Don’t miss your chance to present your work and connect with the community!
See you there!



rdcu.be/eraxZ
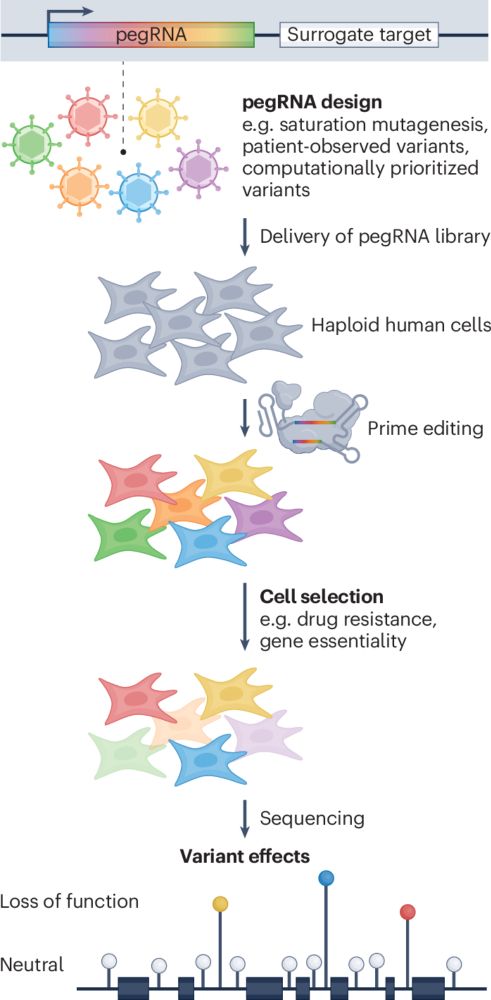
rdcu.be/eraxZ
I'll be sharing insights from my current project on the Discovery Stage at 2 pm.
Come say hi if you're around!

I'll be sharing insights from my current project on the Discovery Stage at 2 pm.
Come say hi if you're around!
You can read all about her work here:
www.cell.com/cell-genomic...
You can read all about her work here:
www.cell.com/cell-genomic...



