
The Genome Function Laboratory
The Francis Crick Institute, London
www.biorxiv.org/content/10.1...
This work was led by Magdalena Armas Reyes, a @crick.ac.uk PhD student until very recently. Congrats, Dr. Armas!
🧵 1/9
We especially encourage students and postdocs to attend and share their work. All talks (except the keynotes) will be selected from abstracts.
I’m delighted to share that registration is now open for the UK Proteostasis Meeting 2026, hosted by The Francis Crick Institute on 20–21 July 2026 .
Please register here(lnkd.in/ervXMzWN) and through Eventbrite for payment (lnkd.in/eTxqjnQy)

We especially encourage students and postdocs to attend and share their work. All talks (except the keynotes) will be selected from abstracts.
www.biorxiv.org/content/10.1...
This work was led by Magdalena Armas Reyes, a @crick.ac.uk PhD student until very recently. Congrats, Dr. Armas!
🧵 1/9
www.biorxiv.org/content/10.1...
This work was led by Magdalena Armas Reyes, a @crick.ac.uk PhD student until very recently. Congrats, Dr. Armas!
🧵 1/9
Henry Scowcroft explores how scientists like @gregfindlay.bsky.social and @carovinuesa.bsky.social are helping unravel the effects of these variants, where even a small change can have a big impact on our lives.
www.crick.ac.uk/news/2025-10...

Henry Scowcroft explores how scientists like @gregfindlay.bsky.social and @carovinuesa.bsky.social are helping unravel the effects of these variants, where even a small change can have a big impact on our lives.
www.crick.ac.uk/news/2025-10...
We're looking for clinicians who are passionate about research to join the 3-year fully funded programme.
Learn more and see what positions are available ⬇️
www.crick.ac.uk/careers-and-...

We're looking for clinicians who are passionate about research to join the 3-year fully funded programme.
Learn more and see what positions are available ⬇️
www.crick.ac.uk/careers-and-...
This is an amalgamation of our two recent preprints - working with @gregfindlay.bsky.social , @cassimons.bsky.social , @dgmacarthur.bsky.social and many others to study variation across RNU4-2 and describe a new recessive NDD 🧬

This is an amalgamation of our two recent preprints - working with @gregfindlay.bsky.social , @cassimons.bsky.social , @dgmacarthur.bsky.social and many others to study variation across RNU4-2 and describe a new recessive NDD 🧬

Check out our talks over the next few days - all unpublished stories.
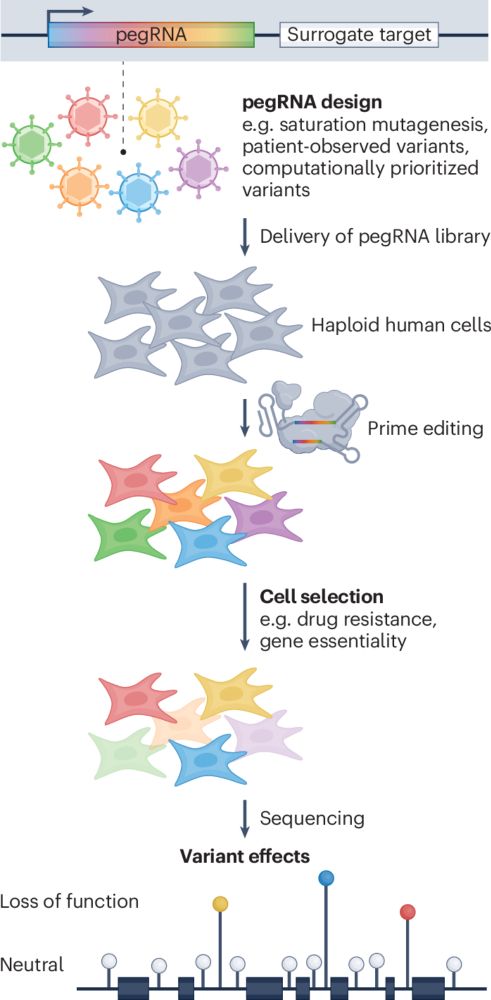
Kicking things off is @michaelherger.bsky.social presenting "Saturation mutagenesis of 37 human splicing factor genes with pooled prime editing". Today @2pm, Rm205abc
Also 👇
Check out our talks over the next few days - all unpublished stories.
Kicking things off is @michaelherger.bsky.social presenting "Saturation mutagenesis of 37 human splicing factor genes with pooled prime editing". Today @2pm, Rm205abc
Also 👇
Hear our Director, Edith Heard, explain why the Crick is a unique place for curiosity-driven research.
Apply now ➡️ www.crick.ac.uk/careers-stud...
We are pleased to be recruiting this year. 👇
www.crick.ac.uk/careers-stud...
We are pleased to be recruiting this year. 👇
www.crick.ac.uk/careers-stud...
Apply by 14 November 2025. 👇
www.crick.ac.uk/careers-stud...

Apply by 14 November 2025. 👇
www.crick.ac.uk/careers-stud...
crick.wd3.myworkdayjobs.com/External/job...
Please apply if you have a background in functional genomics or a related field and are eager to develop methods to map variant effects at scale.
crick.wd3.myworkdayjobs.com/External/job...
Please apply if you have a background in functional genomics or a related field and are eager to develop methods to map variant effects at scale.
www.crick.ac.uk/news/2025-09...

tinyurl.com/3j9r56s8
@rociorius.bsky.social @yuyangchen.bsky.social @gregfindlay.bsky.social @dgmacarthur.bsky.social @cassimons.bsky.social @nickywhiffin.bsky.social

tinyurl.com/3j9r56s8
@rociorius.bsky.social @yuyangchen.bsky.social @gregfindlay.bsky.social @dgmacarthur.bsky.social @cassimons.bsky.social @nickywhiffin.bsky.social

tinyurl.com/3j9r56s8
@rociorius.bsky.social @yuyangchen.bsky.social @gregfindlay.bsky.social @dgmacarthur.bsky.social @cassimons.bsky.social @nickywhiffin.bsky.social
www.medrxiv.org/content/10.1...
Saturation genome editing of BRCA1 across cell types accurately resolves cancer risk.
Led by the amazing Phoebe Dace. This one’s packed full of data, so check out the paper. Quick highlights… 🧵 1/n

www.medrxiv.org/content/10.1...
Saturation genome editing of BRCA1 across cell types accurately resolves cancer risk.
Led by the amazing Phoebe Dace. This one’s packed full of data, so check out the paper. Quick highlights… 🧵 1/n
rdcu.be/eraxZ
rdcu.be/eraxZ
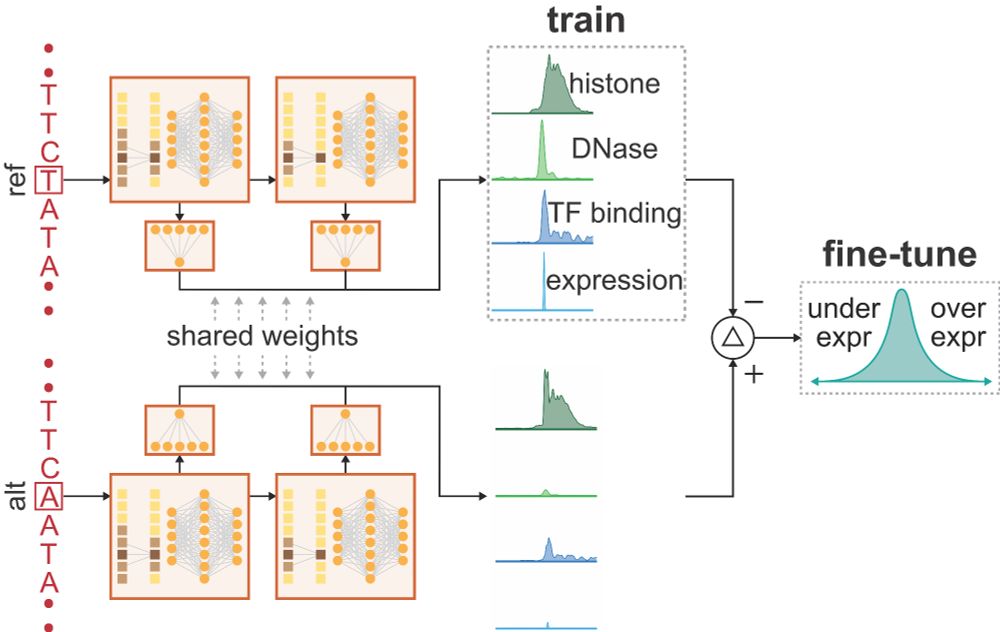
A trio of papers provide key insights for integrating functional data into clinical variant interpretation
#MAVE #VUS
@dougfowler.bsky.social @clareturnbull.bsky.social @leastarita.bsky.social @gregfindlay.bsky.social @afrubin.bsky.social
A trio of papers provide key insights for integrating functional data into clinical variant interpretation
#MAVE #VUS
@dougfowler.bsky.social @clareturnbull.bsky.social @leastarita.bsky.social @gregfindlay.bsky.social @afrubin.bsky.social
You can read all about her work here:
www.cell.com/cell-genomic...
You can read all about her work here:
www.cell.com/cell-genomic...
Highly collaborative project funded by My Name'5 Doddie Foundation
@MNDoddie5
Please repost and share widely!
Highly collaborative project funded by My Name'5 Doddie Foundation
@MNDoddie5
Please repost and share widely!
www.sciencedirect.com/science/arti...

www.sciencedirect.com/science/arti...
Join our team (w/ Prof Hugh Brady @Imperial + Dr Jacob Bush @GSK) on an exciting PhD project developing a covalent cyclic peptide discovery platform with a focus on immuno-oncology targets 🔬🔥
📅 Deadline: 27 April 2025
Join our team (w/ Prof Hugh Brady @Imperial + Dr Jacob Bush @GSK) on an exciting PhD project developing a covalent cyclic peptide discovery platform with a focus on immuno-oncology targets 🔬🔥
📅 Deadline: 27 April 2025
www.medrxiv.org/content/10.1...
A super fun collaboration with incredible duo @gregfindlay.bsky.social @joachimdejonghe.bsky.social from @crick.ac.uk
🧬🖥️🩺
🧵1/12



