us-hupo.org/Distinguishe...

us-hupo.org/Distinguishe...
us-hupo.org/Computationa...

us-hupo.org/Computationa...
www.nature.com/articles/s41...

www.nature.com/articles/s41...
www.nature.com/articles/s41...

www.nature.com/articles/s41...
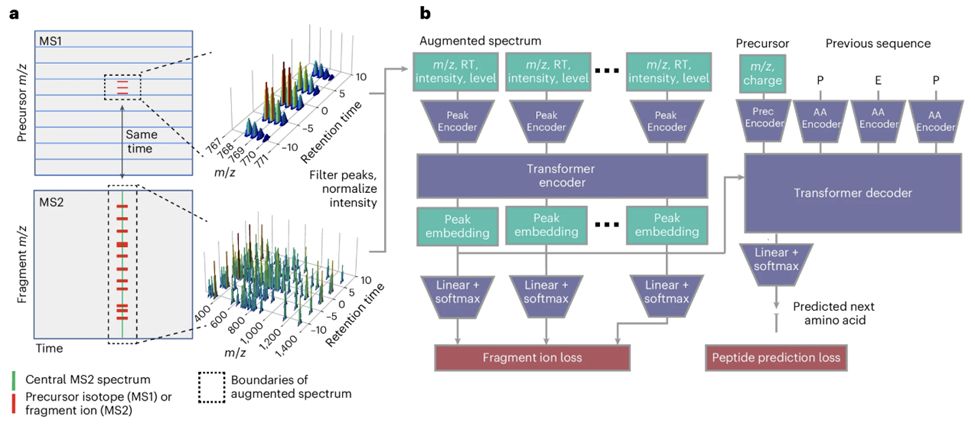

www.nature.com/articles/s41...
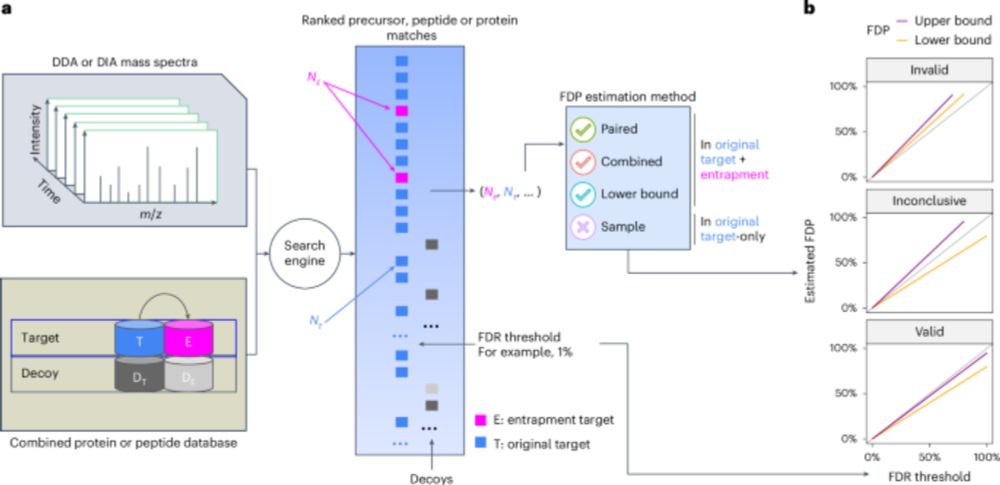
www.nature.com/articles/s41...
www.nature.com/articles/s41...
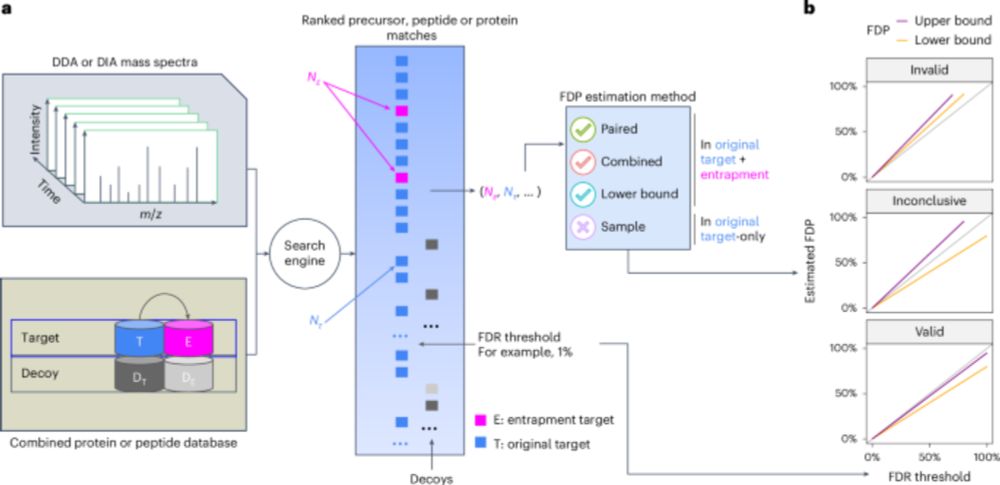
www.nature.com/articles/s41...

www.nature.com/articles/s41...

